Group By: split-apply-combine¶
By “group by” we are referring to a process involving one or more of the following steps
- Splitting the data into groups based on some criteria
- Applying a function to each group independently
- Combining the results into a data structure
Of these, the split step is the most straightforward. In fact, in many situations you may wish to split the data set into groups and do something with those groups yourself. In the apply step, we might wish to one of the following:
Aggregation: computing a summary statistic (or statistics) about each group. Some examples:
- Compute group sums or means
- Compute group sizes / counts
Transformation: perform some group-specific computations and return a like-indexed. Some examples:
- Standardizing data (zscore) within group
- Filling NAs within groups with a value derived from each group
Filtration: discard some groups, according to a group-wise computation that evaluates True or False. Some examples:
- Discarding data that belongs to groups with only a few members
- Filtering out data based on the group sum or mean
Some combination of the above: GroupBy will examine the results of the apply step and try to return a sensibly combined result if it doesn’t fit into either of the above two categories
Since the set of object instance method on pandas data structures are generally rich and expressive, we often simply want to invoke, say, a DataFrame function on each group. The name GroupBy should be quite familiar to those who have used a SQL-based tool (or itertools), in which you can write code like:
SELECT Column1, Column2, mean(Column3), sum(Column4)
FROM SomeTable
GROUP BY Column1, Column2
We aim to make operations like this natural and easy to express using pandas. We’ll address each area of GroupBy functionality then provide some non-trivial examples / use cases.
See the cookbook for some advanced strategies
Splitting an object into groups¶
pandas objects can be split on any of their axes. The abstract definition of grouping is to provide a mapping of labels to group names. To create a GroupBy object (more on what the GroupBy object is later), you do the following:
>>> grouped = obj.groupby(key)
>>> grouped = obj.groupby(key, axis=1)
>>> grouped = obj.groupby([key1, key2])
The mapping can be specified many different ways:
- A Python function, to be called on each of the axis labels
- A list or NumPy array of the same length as the selected axis
- A dict or Series, providing a label -> group name mapping
- For DataFrame objects, a string indicating a column to be used to group. Of course df.groupby('A') is just syntactic sugar for df.groupby(df['A']), but it makes life simpler
- A list of any of the above things
Collectively we refer to the grouping objects as the keys. For example, consider the following DataFrame:
In [1]: df = DataFrame({'A' : ['foo', 'bar', 'foo', 'bar',
...: 'foo', 'bar', 'foo', 'foo'],
...: 'B' : ['one', 'one', 'two', 'three',
...: 'two', 'two', 'one', 'three'],
...: 'C' : randn(8), 'D' : randn(8)})
...:
In [2]: df
Out[2]:
A B C D
0 foo one 0.469112 -0.861849
1 bar one -0.282863 -2.104569
2 foo two -1.509059 -0.494929
3 bar three -1.135632 1.071804
4 foo two 1.212112 0.721555
5 bar two -0.173215 -0.706771
6 foo one 0.119209 -1.039575
7 foo three -1.044236 0.271860
We could naturally group by either the A or B columns or both:
In [3]: grouped = df.groupby('A')
In [4]: grouped = df.groupby(['A', 'B'])
These will split the DataFrame on its index (rows). We could also split by the columns:
In [5]: def get_letter_type(letter):
...: if letter.lower() in 'aeiou':
...: return 'vowel'
...: else:
...: return 'consonant'
...:
In [6]: grouped = df.groupby(get_letter_type, axis=1)
Starting with 0.8, pandas Index objects now supports duplicate values. If a non-unique index is used as the group key in a groupby operation, all values for the same index value will be considered to be in one group and thus the output of aggregation functions will only contain unique index values:
In [7]: lst = [1, 2, 3, 1, 2, 3]
In [8]: s = Series([1, 2, 3, 10, 20, 30], lst)
In [9]: grouped = s.groupby(level=0)
In [10]: grouped.first()
Out[10]:
1 1
2 2
3 3
dtype: int64
In [11]: grouped.last()
Out[11]:
1 10
2 20
3 30
dtype: int64
In [12]: grouped.sum()
Out[12]:
1 11
2 22
3 33
dtype: int64
Note that no splitting occurs until it’s needed. Creating the GroupBy object only verifies that you’ve passed a valid mapping.
Note
Many kinds of complicated data manipulations can be expressed in terms of GroupBy operations (though can’t be guaranteed to be the most efficient). You can get quite creative with the label mapping functions.
GroupBy object attributes¶
The groups attribute is a dict whose keys are the computed unique groups and corresponding values being the axis labels belonging to each group. In the above example we have:
In [13]: df.groupby('A').groups
Out[13]: {'bar': [1L, 3L, 5L], 'foo': [0L, 2L, 4L, 6L, 7L]}
In [14]: df.groupby(get_letter_type, axis=1).groups
Out[14]: {'consonant': ['B', 'C', 'D'], 'vowel': ['A']}
Calling the standard Python len function on the GroupBy object just returns the length of the groups dict, so it is largely just a convenience:
In [15]: grouped = df.groupby(['A', 'B'])
In [16]: grouped.groups
Out[16]:
{('bar', 'one'): [1L],
('bar', 'three'): [3L],
('bar', 'two'): [5L],
('foo', 'one'): [0L, 6L],
('foo', 'three'): [7L],
('foo', 'two'): [2L, 4L]}
In [17]: len(grouped)
Out[17]: 6
By default the group keys are sorted during the groupby operation. You may however pass sort=False for potential speedups:
In [18]: df2 = DataFrame({'X' : ['B', 'B', 'A', 'A'], 'Y' : [1, 2, 3, 4]})
In [19]: df2.groupby(['X'], sort=True).sum()
Out[19]:
Y
X
A 7
B 3
In [20]: df2.groupby(['X'], sort=False).sum()
Out[20]:
Y
X
B 3
A 7
GroupBy will tab complete column names (and other attributes)
In [21]: df
Out[21]:
gender height weight
2000-01-01 male 42.849980 157.500553
2000-01-02 male 49.607315 177.340407
2000-01-03 male 56.293531 171.524640
2000-01-04 female 48.421077 144.251986
2000-01-05 male 46.556882 152.526206
2000-01-06 female 68.448851 168.272968
2000-01-07 male 70.757698 136.431469
2000-01-08 female 58.909500 176.499753
2000-01-09 female 76.435631 174.094104
2000-01-10 male 45.306120 177.540920
In [22]: gb = df.groupby('gender')
In [23]: gb.<TAB>
gb.agg gb.boxplot gb.cummin gb.describe gb.filter gb.get_group gb.height gb.last gb.median gb.ngroups gb.plot gb.rank gb.std gb.transform
gb.aggregate gb.count gb.cumprod gb.dtype gb.first gb.groups gb.hist gb.max gb.min gb.nth gb.prod gb.resample gb.sum gb.var
gb.apply gb.cummax gb.cumsum gb.fillna gb.gender gb.head gb.indices gb.mean gb.name gb.ohlc gb.quantile gb.size gb.tail gb.weight
GroupBy with MultiIndex¶
With hierarchically-indexed data, it’s quite natural to group by one of the levels of the hierarchy.
In [24]: s
Out[24]:
first second
bar one -0.575247
two 0.254161
baz one -1.143704
two 0.215897
foo one 1.193555
two -0.077118
qux one -0.408530
two -0.862495
dtype: float64
In [25]: grouped = s.groupby(level=0)
In [26]: grouped.sum()
Out[26]:
first
bar -0.321085
baz -0.927807
foo 1.116437
qux -1.271025
dtype: float64
If the MultiIndex has names specified, these can be passed instead of the level number:
In [27]: s.groupby(level='second').sum()
Out[27]:
second
one -0.933926
two -0.469555
dtype: float64
The aggregation functions such as sum will take the level parameter directly. Additionally, the resulting index will be named according to the chosen level:
In [28]: s.sum(level='second')
Out[28]:
second
one -0.933926
two -0.469555
dtype: float64
Also as of v0.6, grouping with multiple levels is supported.
In [29]: s
Out[29]:
first second third
bar doo one 1.346061
two 1.511763
baz bee one 1.627081
two -0.990582
foo bop one -0.441652
two 1.211526
qux bop one 0.268520
two 0.024580
dtype: float64
In [30]: s.groupby(level=['first','second']).sum()
Out[30]:
first second
bar doo 2.857824
baz bee 0.636499
foo bop 0.769873
qux bop 0.293100
dtype: float64
More on the sum function and aggregation later.
DataFrame column selection in GroupBy¶
Once you have created the GroupBy object from a DataFrame, for example, you might want to do something different for each of the columns. Thus, using [] similar to getting a column from a DataFrame, you can do:
In [31]: grouped = df.groupby(['A'])
In [32]: grouped_C = grouped['C']
In [33]: grouped_D = grouped['D']
This is mainly syntactic sugar for the alternative and much more verbose:
In [34]: df['C'].groupby(df['A'])
Out[34]: <pandas.core.groupby.SeriesGroupBy object at 0xad5eefac>
Additionally this method avoids recomputing the internal grouping information derived from the passed key.
Iterating through groups¶
With the GroupBy object in hand, iterating through the grouped data is very natural and functions similarly to itertools.groupby:
In [35]: grouped = df.groupby('A')
In [36]: for name, group in grouped:
....: print(name)
....: print(group)
....:
bar
A B C D
1 bar one -0.042379 -0.089329
3 bar three -0.009920 -0.945867
5 bar two 0.495767 1.956030
foo
A B C D
0 foo one -0.919854 -1.131345
2 foo two 1.247642 0.337863
4 foo two 0.290213 -0.932132
6 foo one 0.362949 0.017587
7 foo three 1.548106 -0.016692
In the case of grouping by multiple keys, the group name will be a tuple:
In [37]: for name, group in df.groupby(['A', 'B']):
....: print(name)
....: print(group)
....:
('bar', 'one')
A B C D
1 bar one -0.042379 -0.089329
('bar', 'three')
A B C D
3 bar three -0.00992 -0.945867
('bar', 'two')
A B C D
5 bar two 0.495767 1.95603
('foo', 'one')
A B C D
0 foo one -0.919854 -1.131345
6 foo one 0.362949 0.017587
('foo', 'three')
A B C D
7 foo three 1.548106 -0.016692
('foo', 'two')
A B C D
2 foo two 1.247642 0.337863
4 foo two 0.290213 -0.932132
It’s standard Python-fu but remember you can unpack the tuple in the for loop statement if you wish: for (k1, k2), group in grouped:.
Aggregation¶
Once the GroupBy object has been created, several methods are available to perform a computation on the grouped data.
An obvious one is aggregation via the aggregate or equivalently agg method:
In [38]: grouped = df.groupby('A')
In [39]: grouped.aggregate(np.sum)
Out[39]:
C D
A
bar 0.443469 0.920834
foo 2.529056 -1.724719
In [40]: grouped = df.groupby(['A', 'B'])
In [41]: grouped.aggregate(np.sum)
Out[41]:
C D
A B
bar one -0.042379 -0.089329
three -0.009920 -0.945867
two 0.495767 1.956030
foo one -0.556905 -1.113758
three 1.548106 -0.016692
two 1.537855 -0.594269
As you can see, the result of the aggregation will have the group names as the new index along the grouped axis. In the case of multiple keys, the result is a MultiIndex by default, though this can be changed by using the as_index option:
In [42]: grouped = df.groupby(['A', 'B'], as_index=False)
In [43]: grouped.aggregate(np.sum)
Out[43]:
A B C D
0 bar one -0.042379 -0.089329
1 bar three -0.009920 -0.945867
2 bar two 0.495767 1.956030
3 foo one -0.556905 -1.113758
4 foo three 1.548106 -0.016692
5 foo two 1.537855 -0.594269
In [44]: df.groupby('A', as_index=False).sum()
Out[44]:
A C D
0 bar 0.443469 0.920834
1 foo 2.529056 -1.724719
Note that you could use the reset_index DataFrame function to achieve the same result as the column names are stored in the resulting MultiIndex:
In [45]: df.groupby(['A', 'B']).sum().reset_index()
Out[45]:
A B C D
0 bar one -0.042379 -0.089329
1 bar three -0.009920 -0.945867
2 bar two 0.495767 1.956030
3 foo one -0.556905 -1.113758
4 foo three 1.548106 -0.016692
5 foo two 1.537855 -0.594269
Another simple aggregation example is to compute the size of each group. This is included in GroupBy as the size method. It returns a Series whose index are the group names and whose values are the sizes of each group.
In [46]: grouped.size()
Out[46]:
A B
bar one 1
three 1
two 1
foo one 2
three 1
two 2
dtype: int64
In [47]: grouped.describe()
Out[47]:
C D
0 count 1.000000 1.000000
mean -0.042379 -0.089329
std NaN NaN
min -0.042379 -0.089329
25% -0.042379 -0.089329
50% -0.042379 -0.089329
75% -0.042379 -0.089329
... ... ...
5 mean 0.768928 -0.297134
std 0.677005 0.898022
min 0.290213 -0.932132
25% 0.529570 -0.614633
50% 0.768928 -0.297134
75% 1.008285 0.020364
max 1.247642 0.337863
[48 rows x 2 columns]
Note
Aggregation functions will not return the groups that you are aggregating over if they are named columns, when as_index=True, the default. The grouped columns will be the indices of the returned object.
Passing as_index=False will return the groups that you are aggregating over, if they are named columns.
Aggregating functions are ones that reduce the dimension of the returned objects, for example: mean, sum, size, count, std, var, sem, describe, first, last, nth, min, max. This is what happens when you do for example DataFrame.sum() and get back a Series.
nth can act as a reducer or a filter, see here
Applying multiple functions at once¶
With grouped Series you can also pass a list or dict of functions to do aggregation with, outputting a DataFrame:
In [48]: grouped = df.groupby('A')
In [49]: grouped['C'].agg([np.sum, np.mean, np.std])
Out[49]:
sum mean std
A
bar 0.443469 0.147823 0.301765
foo 2.529056 0.505811 0.966450
If a dict is passed, the keys will be used to name the columns. Otherwise the function’s name (stored in the function object) will be used.
In [50]: grouped['D'].agg({'result1' : np.sum,
....: 'result2' : np.mean})
....:
Out[50]:
result2 result1
A
bar 0.306945 0.920834
foo -0.344944 -1.724719
On a grouped DataFrame, you can pass a list of functions to apply to each column, which produces an aggregated result with a hierarchical index:
In [51]: grouped.agg([np.sum, np.mean, np.std])
Out[51]:
C D
sum mean std sum mean std
A
bar 0.443469 0.147823 0.301765 0.920834 0.306945 1.490982
foo 2.529056 0.505811 0.966450 -1.724719 -0.344944 0.645875
Passing a dict of functions has different behavior by default, see the next section.
Applying different functions to DataFrame columns¶
By passing a dict to aggregate you can apply a different aggregation to the columns of a DataFrame:
In [52]: grouped.agg({'C' : np.sum,
....: 'D' : lambda x: np.std(x, ddof=1)})
....:
Out[52]:
C D
A
bar 0.443469 1.490982
foo 2.529056 0.645875
The function names can also be strings. In order for a string to be valid it must be either implemented on GroupBy or available via dispatching:
In [53]: grouped.agg({'C' : 'sum', 'D' : 'std'})
Out[53]:
C D
A
bar 0.443469 1.490982
foo 2.529056 0.645875
Cython-optimized aggregation functions¶
Some common aggregations, currently only sum, mean, std, and sem, have optimized Cython implementations:
In [54]: df.groupby('A').sum()
Out[54]:
C D
A
bar 0.443469 0.920834
foo 2.529056 -1.724719
In [55]: df.groupby(['A', 'B']).mean()
Out[55]:
C D
A B
bar one -0.042379 -0.089329
three -0.009920 -0.945867
two 0.495767 1.956030
foo one -0.278452 -0.556879
three 1.548106 -0.016692
two 0.768928 -0.297134
Of course sum and mean are implemented on pandas objects, so the above code would work even without the special versions via dispatching (see below).
Transformation¶
The transform method returns an object that is indexed the same (same size) as the one being grouped. Thus, the passed transform function should return a result that is the same size as the group chunk. For example, suppose we wished to standardize the data within each group:
In [56]: index = date_range('10/1/1999', periods=1100)
In [57]: ts = Series(np.random.normal(0.5, 2, 1100), index)
In [58]: ts = rolling_mean(ts, 100, 100).dropna()
In [59]: ts.head()
Out[59]:
2000-01-08 0.779333
2000-01-09 0.778852
2000-01-10 0.786476
2000-01-11 0.782797
2000-01-12 0.798110
Freq: D, dtype: float64
In [60]: ts.tail()
Out[60]:
2002-09-30 0.660294
2002-10-01 0.631095
2002-10-02 0.673601
2002-10-03 0.709213
2002-10-04 0.719369
Freq: D, dtype: float64
In [61]: key = lambda x: x.year
In [62]: zscore = lambda x: (x - x.mean()) / x.std()
In [63]: transformed = ts.groupby(key).transform(zscore)
We would expect the result to now have mean 0 and standard deviation 1 within each group, which we can easily check:
# Original Data
In [64]: grouped = ts.groupby(key)
In [65]: grouped.mean()
Out[65]:
2000 0.442441
2001 0.526246
2002 0.459365
dtype: float64
In [66]: grouped.std()
Out[66]:
2000 0.131752
2001 0.210945
2002 0.128753
dtype: float64
# Transformed Data
In [67]: grouped_trans = transformed.groupby(key)
In [68]: grouped_trans.mean()
Out[68]:
2000 -7.561268e-17
2001 -4.194514e-16
2002 -1.362729e-16
dtype: float64
In [69]: grouped_trans.std()
Out[69]:
2000 1
2001 1
2002 1
dtype: float64
We can also visually compare the original and transformed data sets.
In [70]: compare = DataFrame({'Original': ts, 'Transformed': transformed})
In [71]: compare.plot()
Out[71]: <matplotlib.axes._subplots.AxesSubplot at 0xa1807b2c>
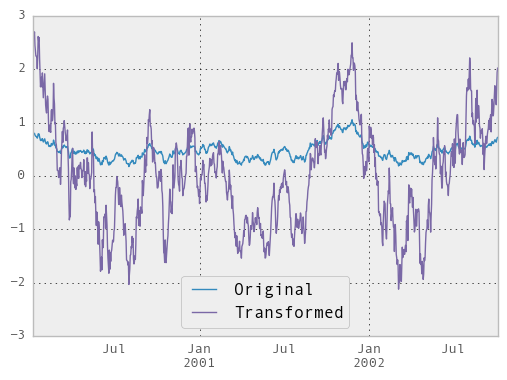
Another common data transform is to replace missing data with the group mean.
In [72]: data_df
Out[72]:
A B C
0 1.539708 -1.166480 0.533026
1 1.302092 -0.505754 NaN
2 -0.371983 1.104803 -0.651520
3 -1.309622 1.118697 -1.161657
4 -1.924296 0.396437 0.812436
5 0.815643 0.367816 -0.469478
6 -0.030651 1.376106 -0.645129
.. ... ... ...
993 0.012359 0.554602 -1.976159
994 0.042312 -1.628835 1.013822
995 -0.093110 0.683847 -0.774753
996 -0.185043 1.438572 NaN
997 -0.394469 -0.642343 0.011374
998 -1.174126 1.857148 NaN
999 0.234564 0.517098 0.393534
[1000 rows x 3 columns]
In [73]: countries = np.array(['US', 'UK', 'GR', 'JP'])
In [74]: key = countries[np.random.randint(0, 4, 1000)]
In [75]: grouped = data_df.groupby(key)
# Non-NA count in each group
In [76]: grouped.count()
Out[76]:
A B C
GR 209 217 189
JP 240 255 217
UK 216 231 193
US 239 250 217
In [77]: f = lambda x: x.fillna(x.mean())
In [78]: transformed = grouped.transform(f)
We can verify that the group means have not changed in the transformed data and that the transformed data contains no NAs.
In [79]: grouped_trans = transformed.groupby(key)
In [80]: grouped.mean() # original group means
Out[80]:
A B C
GR -0.098371 -0.015420 0.068053
JP 0.069025 0.023100 -0.077324
UK 0.034069 -0.052580 -0.116525
US 0.058664 -0.020399 0.028603
In [81]: grouped_trans.mean() # transformation did not change group means
Out[81]:
A B C
GR -0.098371 -0.015420 0.068053
JP 0.069025 0.023100 -0.077324
UK 0.034069 -0.052580 -0.116525
US 0.058664 -0.020399 0.028603
In [82]: grouped.count() # original has some missing data points
Out[82]:
A B C
GR 209 217 189
JP 240 255 217
UK 216 231 193
US 239 250 217
In [83]: grouped_trans.count() # counts after transformation
Out[83]:
A B C
GR 228 228 228
JP 267 267 267
UK 247 247 247
US 258 258 258
In [84]: grouped_trans.size() # Verify non-NA count equals group size
Out[84]:
GR 228
JP 267
UK 247
US 258
dtype: int64
Note
Some functions when applied to a groupby object will automatically transform the input, returning an object of the same shape as the original. Passing as_index=False will not affect these transformation methods.
For example: fillna, ffill, bfill, shift.
In [85]: grouped.ffill()
Out[85]:
A B C
0 1.539708 -1.166480 0.533026
1 1.302092 -0.505754 0.533026
2 -0.371983 1.104803 -0.651520
3 -1.309622 1.118697 -1.161657
4 -1.924296 0.396437 0.812436
5 0.815643 0.367816 -0.469478
6 -0.030651 1.376106 -0.645129
.. ... ... ...
993 0.012359 0.554602 -1.976159
994 0.042312 -1.628835 1.013822
995 -0.093110 0.683847 -0.774753
996 -0.185043 1.438572 -0.774753
997 -0.394469 -0.642343 0.011374
998 -1.174126 1.857148 -0.774753
999 0.234564 0.517098 0.393534
[1000 rows x 3 columns]
Filtration¶
New in version 0.12.
The filter method returns a subset of the original object. Suppose we want to take only elements that belong to groups with a group sum greater than 2.
In [86]: sf = Series([1, 1, 2, 3, 3, 3])
In [87]: sf.groupby(sf).filter(lambda x: x.sum() > 2)
Out[87]:
3 3
4 3
5 3
dtype: int64
The argument of filter must be a function that, applied to the group as a whole, returns True or False.
Another useful operation is filtering out elements that belong to groups with only a couple members.
In [88]: dff = DataFrame({'A': np.arange(8), 'B': list('aabbbbcc')})
In [89]: dff.groupby('B').filter(lambda x: len(x) > 2)
Out[89]:
A B
2 2 b
3 3 b
4 4 b
5 5 b
Alternatively, instead of dropping the offending groups, we can return a like-indexed objects where the groups that do not pass the filter are filled with NaNs.
In [90]: dff.groupby('B').filter(lambda x: len(x) > 2, dropna=False)
Out[90]:
A B
0 NaN NaN
1 NaN NaN
2 2 b
3 3 b
4 4 b
5 5 b
6 NaN NaN
7 NaN NaN
For dataframes with multiple columns, filters should explicitly specify a column as the filter criterion.
In [91]: dff['C'] = np.arange(8)
In [92]: dff.groupby('B').filter(lambda x: len(x['C']) > 2)
Out[92]:
A B C
2 2 b 2
3 3 b 3
4 4 b 4
5 5 b 5
Note
Some functions when applied to a groupby object will act as a filter on the input, returning a reduced shape of the original (and potentitally eliminating groups), but with the index unchanged. Passing as_index=False will not affect these transformation methods.
For example: head, tail.
In [93]: dff.groupby('B').head(2)
Out[93]:
A B C
0 0 a 0
1 1 a 1
2 2 b 2
3 3 b 3
6 6 c 6
7 7 c 7
Dispatching to instance methods¶
When doing an aggregation or transformation, you might just want to call an instance method on each data group. This is pretty easy to do by passing lambda functions:
In [94]: grouped = df.groupby('A')
In [95]: grouped.agg(lambda x: x.std())
Out[95]:
B C D
A
bar NaN 0.301765 1.490982
foo NaN 0.966450 0.645875
But, it’s rather verbose and can be untidy if you need to pass additional arguments. Using a bit of metaprogramming cleverness, GroupBy now has the ability to “dispatch” method calls to the groups:
In [96]: grouped.std()
Out[96]:
C D
A
bar 0.301765 1.490982
foo 0.966450 0.645875
What is actually happening here is that a function wrapper is being generated. When invoked, it takes any passed arguments and invokes the function with any arguments on each group (in the above example, the std function). The results are then combined together much in the style of agg and transform (it actually uses apply to infer the gluing, documented next). This enables some operations to be carried out rather succinctly:
In [97]: tsdf = DataFrame(randn(1000, 3),
....: index=date_range('1/1/2000', periods=1000),
....: columns=['A', 'B', 'C'])
....:
In [98]: tsdf.ix[::2] = np.nan
In [99]: grouped = tsdf.groupby(lambda x: x.year)
In [100]: grouped.fillna(method='pad')
Out[100]:
A B C
2000-01-01 NaN NaN NaN
2000-01-02 -0.353501 -0.080957 -0.876864
2000-01-03 -0.353501 -0.080957 -0.876864
2000-01-04 0.050976 0.044273 -0.559849
2000-01-05 0.050976 0.044273 -0.559849
2000-01-06 0.030091 0.186460 -0.680149
2000-01-07 0.030091 0.186460 -0.680149
... ... ... ...
2002-09-20 2.310215 0.157482 -0.064476
2002-09-21 2.310215 0.157482 -0.064476
2002-09-22 0.005011 0.053897 -1.026922
2002-09-23 0.005011 0.053897 -1.026922
2002-09-24 -0.456542 -1.849051 1.559856
2002-09-25 -0.456542 -1.849051 1.559856
2002-09-26 1.123162 0.354660 1.128135
[1000 rows x 3 columns]
In this example, we chopped the collection of time series into yearly chunks then independently called fillna on the groups.
New in version 0.14.1.
The nlargest and nsmallest methods work on Series style groupbys:
In [101]: s = Series([9, 8, 7, 5, 19, 1, 4.2, 3.3])
In [102]: g = Series(list('abababab'))
In [103]: gb = s.groupby(g)
In [104]: gb.nlargest(3)
Out[104]:
a 4 19.0
0 9.0
2 7.0
b 1 8.0
3 5.0
7 3.3
dtype: float64
In [105]: gb.nsmallest(3)
Out[105]:
a 6 4.2
2 7.0
0 9.0
b 5 1.0
7 3.3
3 5.0
dtype: float64
Flexible apply¶
Some operations on the grouped data might not fit into either the aggregate or transform categories. Or, you may simply want GroupBy to infer how to combine the results. For these, use the apply function, which can be substituted for both aggregate and transform in many standard use cases. However, apply can handle some exceptional use cases, for example:
In [106]: df
Out[106]:
A B C D
0 foo one -0.919854 -1.131345
1 bar one -0.042379 -0.089329
2 foo two 1.247642 0.337863
3 bar three -0.009920 -0.945867
4 foo two 0.290213 -0.932132
5 bar two 0.495767 1.956030
6 foo one 0.362949 0.017587
7 foo three 1.548106 -0.016692
In [107]: grouped = df.groupby('A')
# could also just call .describe()
In [108]: grouped['C'].apply(lambda x: x.describe())
Out[108]:
A
bar count 3.000000
mean 0.147823
std 0.301765
min -0.042379
25% -0.026149
...
foo std 0.966450
min -0.919854
25% 0.290213
50% 0.362949
75% 1.247642
max 1.548106
Length: 16, dtype: float64
The dimension of the returned result can also change:
In [109]: grouped = df.groupby('A')['C']
In [110]: def f(group):
.....: return DataFrame({'original' : group,
.....: 'demeaned' : group - group.mean()})
.....:
In [111]: grouped.apply(f)
Out[111]:
demeaned original
0 -1.425665 -0.919854
1 -0.190202 -0.042379
2 0.741831 1.247642
3 -0.157743 -0.009920
4 -0.215598 0.290213
5 0.347944 0.495767
6 -0.142862 0.362949
7 1.042295 1.548106
apply on a Series can operate on a returned value from the applied function, that is itself a series, and possibly upcast the result to a DataFrame
In [112]: def f(x):
.....: return Series([ x, x**2 ], index = ['x', 'x^s'])
.....:
In [113]: s
Out[113]:
0 9.0
1 8.0
2 7.0
3 5.0
4 19.0
5 1.0
6 4.2
7 3.3
dtype: float64
In [114]: s.apply(f)
Out[114]:
x x^s
0 9.0 81.00
1 8.0 64.00
2 7.0 49.00
3 5.0 25.00
4 19.0 361.00
5 1.0 1.00
6 4.2 17.64
7 3.3 10.89
Note
apply can act as a reducer, transformer, or filter function, depending on exactly what is passed to apply. So depending on the path taken, and exactly what you are grouping. Thus the grouped columns(s) may be included in the output as well as set the indices.
Warning
In the current implementation apply calls func twice on the first group to decide whether it can take a fast or slow code path. This can lead to unexpected behavior if func has side-effects, as they will take effect twice for the first group.
In [115]: d = DataFrame({"a":["x", "y"], "b":[1,2]})
In [116]: def identity(df):
.....: print df
.....: return df
.....:
In [117]: d.groupby("a").apply(identity)
a b
0 x 1
a b
0 x 1
a b
1 y 2
Out[117]:
a b
0 x 1
1 y 2
Other useful features¶
Automatic exclusion of “nuisance” columns¶
Again consider the example DataFrame we’ve been looking at:
In [118]: df
Out[118]:
A B C D
0 foo one -0.919854 -1.131345
1 bar one -0.042379 -0.089329
2 foo two 1.247642 0.337863
3 bar three -0.009920 -0.945867
4 foo two 0.290213 -0.932132
5 bar two 0.495767 1.956030
6 foo one 0.362949 0.017587
7 foo three 1.548106 -0.016692
Supposed we wished to compute the standard deviation grouped by the A column. There is a slight problem, namely that we don’t care about the data in column B. We refer to this as a “nuisance” column. If the passed aggregation function can’t be applied to some columns, the troublesome columns will be (silently) dropped. Thus, this does not pose any problems:
In [119]: df.groupby('A').std()
Out[119]:
C D
A
bar 0.301765 1.490982
foo 0.966450 0.645875
NA group handling¶
If there are any NaN values in the grouping key, these will be automatically excluded. So there will never be an “NA group”. This was not the case in older versions of pandas, but users were generally discarding the NA group anyway (and supporting it was an implementation headache).
Grouping with ordered factors¶
Categorical variables represented as instance of pandas’s Categorical class can be used as group keys. If so, the order of the levels will be preserved:
In [120]: data = Series(np.random.randn(100))
In [121]: factor = qcut(data, [0, .25, .5, .75, 1.])
In [122]: data.groupby(factor).mean()
Out[122]:
[-2.617, -0.684] -1.331461
(-0.684, -0.0232] -0.272816
(-0.0232, 0.541] 0.263607
(0.541, 2.369] 1.166038
dtype: float64
Grouping with a Grouper specification¶
Your may need to specify a bit more data to properly group. You can use the pd.Grouper to provide this local control.
In [123]: import datetime as DT
In [124]: df = DataFrame({
.....: 'Branch' : 'A A A A A A A B'.split(),
.....: 'Buyer': 'Carl Mark Carl Carl Joe Joe Joe Carl'.split(),
.....: 'Quantity': [1,3,5,1,8,1,9,3],
.....: 'Date' : [
.....: DT.datetime(2013,1,1,13,0),
.....: DT.datetime(2013,1,1,13,5),
.....: DT.datetime(2013,10,1,20,0),
.....: DT.datetime(2013,10,2,10,0),
.....: DT.datetime(2013,10,1,20,0),
.....: DT.datetime(2013,10,2,10,0),
.....: DT.datetime(2013,12,2,12,0),
.....: DT.datetime(2013,12,2,14,0),
.....: ]})
.....:
In [125]: df
Out[125]:
Branch Buyer Date Quantity
0 A Carl 2013-01-01 13:00:00 1
1 A Mark 2013-01-01 13:05:00 3
2 A Carl 2013-10-01 20:00:00 5
3 A Carl 2013-10-02 10:00:00 1
4 A Joe 2013-10-01 20:00:00 8
5 A Joe 2013-10-02 10:00:00 1
6 A Joe 2013-12-02 12:00:00 9
7 B Carl 2013-12-02 14:00:00 3
Groupby a specific column with the desired frequency. This is like resampling.
In [126]: df.groupby([pd.Grouper(freq='1M',key='Date'),'Buyer']).sum()
Out[126]:
Quantity
Date Buyer
2013-01-31 Carl 1
Mark 3
2013-10-31 Carl 6
Joe 9
2013-12-31 Carl 3
Joe 9
You have an ambiguous specification in that you have a named index and a column that could be potential groupers.
In [127]: df = df.set_index('Date')
In [128]: df['Date'] = df.index + pd.offsets.MonthEnd(2)
In [129]: df.groupby([pd.Grouper(freq='6M',key='Date'),'Buyer']).sum()
Out[129]:
Quantity
Date Buyer
2013-02-28 Carl 1
Mark 3
2014-02-28 Carl 9
Joe 18
In [130]: df.groupby([pd.Grouper(freq='6M',level='Date'),'Buyer']).sum()
Out[130]:
Quantity
Date Buyer
2013-01-31 Carl 1
Mark 3
2014-01-31 Carl 9
Joe 18
Taking the first rows of each group¶
Just like for a DataFrame or Series you can call head and tail on a groupby:
In [131]: df = DataFrame([[1, 2], [1, 4], [5, 6]], columns=['A', 'B'])
In [132]: df
Out[132]:
A B
0 1 2
1 1 4
2 5 6
In [133]: g = df.groupby('A')
In [134]: g.head(1)
Out[134]:
A B
0 1 2
2 5 6
In [135]: g.tail(1)
Out[135]:
A B
1 1 4
2 5 6
This shows the first or last n rows from each group.
Warning
Before 0.14.0 this was implemented with a fall-through apply, so the result would incorrectly respect the as_index flag:
>>> g.head(1): # was equivalent to g.apply(lambda x: x.head(1))
A B
A
1 0 1 2
5 2 5 6
Taking the nth row of each group¶
To select from a DataFrame or Series the nth item, use the nth method. This is a reduction method, and will return a single row (or no row) per group if you pass an int for n:
In [136]: df = DataFrame([[1, np.nan], [1, 4], [5, 6]], columns=['A', 'B'])
In [137]: g = df.groupby('A')
In [138]: g.nth(0)
Out[138]:
B
A
1 NaN
5 6
In [139]: g.nth(-1)
Out[139]:
B
A
1 4
5 6
In [140]: g.nth(1)
Out[140]:
B
A
1 4
If you want to select the nth not-null item, use the dropna kwarg. For a DataFrame this should be either 'any' or 'all' just like you would pass to dropna, for a Series this just needs to be truthy.
# nth(0) is the same as g.first()
In [141]: g.nth(0, dropna='any')
Out[141]:
B
A
1 4
5 6
In [142]: g.first()
Out[142]:
B
A
1 4
5 6
# nth(-1) is the same as g.last()
In [143]: g.nth(-1, dropna='any') # NaNs denote group exhausted when using dropna
Out[143]:
B
A
1 4
5 6
In [144]: g.last()
Out[144]:
B
A
1 4
5 6
In [145]: g.B.nth(0, dropna=True)
Out[145]:
A
1 4
5 6
Name: B, dtype: float64
As with other methods, passing as_index=False, will achieve a filtration, which returns the grouped row.
In [146]: df = DataFrame([[1, np.nan], [1, 4], [5, 6]], columns=['A', 'B'])
In [147]: g = df.groupby('A',as_index=False)
In [148]: g.nth(0)
Out[148]:
A B
0 1 NaN
2 5 6
In [149]: g.nth(-1)
Out[149]:
A B
1 1 4
2 5 6
You can also select multiple rows from each group by specifying multiple nth values as a list of ints.
In [150]: business_dates = date_range(start='4/1/2014', end='6/30/2014', freq='B')
In [151]: df = DataFrame(1, index=business_dates, columns=['a', 'b'])
# get the first, 4th, and last date index for each month
In [152]: df.groupby((df.index.year, df.index.month)).nth([0, 3, -1])
Out[152]:
a b
2014-04-01 1 1
2014-04-04 1 1
2014-04-30 1 1
2014-05-01 1 1
2014-05-06 1 1
2014-05-30 1 1
2014-06-02 1 1
2014-06-05 1 1
2014-06-30 1 1
Enumerate group items¶
New in version 0.13.0.
To see the order in which each row appears within its group, use the cumcount method:
In [153]: df = pd.DataFrame(list('aaabba'), columns=['A'])
In [154]: df
Out[154]:
A
0 a
1 a
2 a
3 b
4 b
5 a
In [155]: df.groupby('A').cumcount()
Out[155]:
0 0
1 1
2 2
3 0
4 1
5 3
dtype: int64
In [156]: df.groupby('A').cumcount(ascending=False) # kwarg only
Out[156]:
0 3
1 2
2 1
3 1
4 0
5 0
dtype: int64
Plotting¶
Groupby also works with some plotting methods. For example, suppose we suspect that some features in a DataFrame my differ by group, in this case, the values in column 1 where the group is “B” are 3 higher on average.
In [157]: np.random.seed(1234)
In [158]: df = DataFrame(np.random.randn(50, 2))
In [159]: df['g'] = np.random.choice(['A', 'B'], size=50)
In [160]: df.loc[df['g'] == 'B', 1] += 3
We can easily visualize this with a boxplot:
In [161]: df.groupby('g').boxplot()
Out[161]: OrderedDict([('A', {'boxes': [<matplotlib.lines.Line2D object at 0xa14c6a6c>, <matplotlib.lines.Line2D object at 0xa118f72c>], 'fliers': [<matplotlib.lines.Line2D object at 0xa121760c>, <matplotlib.lines.Line2D object at 0xa17fad6c>], 'medians': [<matplotlib.lines.Line2D object at 0xa11a844c>, <matplotlib.lines.Line2D object at 0xa17faecc>], 'means': [], 'whiskers': [<matplotlib.lines.Line2D object at 0xa14f30ac>, <matplotlib.lines.Line2D object at 0xa11968ec>, <matplotlib.lines.Line2D object at 0xa11a972c>, <matplotlib.lines.Line2D object at 0xa12412ac>], 'caps': [<matplotlib.lines.Line2D object at 0xa123e26c>, <matplotlib.lines.Line2D object at 0xa11868ec>, <matplotlib.lines.Line2D object at 0xa11dec4c>, <matplotlib.lines.Line2D object at 0xa11eccec>]}), ('B', {'boxes': [<matplotlib.lines.Line2D object at 0xa17f124c>, <matplotlib.lines.Line2D object at 0xa180096c>], 'fliers': [<matplotlib.lines.Line2D object at 0xa1800ccc>, <matplotlib.lines.Line2D object at 0xa111672c>], 'medians': [<matplotlib.lines.Line2D object at 0xa180056c>, <matplotlib.lines.Line2D object at 0xa1116d2c>], 'means': [], 'whiskers': [<matplotlib.lines.Line2D object at 0xa17f1d6c>, <matplotlib.lines.Line2D object at 0xa17f19ac>, <matplotlib.lines.Line2D object at 0xa17bccac>, <matplotlib.lines.Line2D object at 0xa17bce8c>], 'caps': [<matplotlib.lines.Line2D object at 0xa17f134c>, <matplotlib.lines.Line2D object at 0xa17f1a0c>, <matplotlib.lines.Line2D object at 0xa120b58c>, <matplotlib.lines.Line2D object at 0xa10b476c>]})])
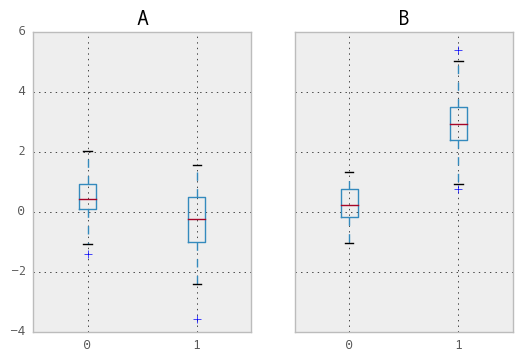
The result of calling boxplot is a dictionary whose keys are the values of our grouping column g (“A” and “B”). The values of the resulting dictionary can be controlled by the return_type keyword of boxplot. See the visualization documentation for more.
Warning
For historical reasons, df.groupby("g").boxplot() is not equivalent to df.boxplot(by="g"). See here for an explanation.
Examples¶
Regrouping by factor¶
Regroup columns of a DataFrame according to their sum, and sum the aggregated ones.
In [162]: df = pd.DataFrame({'a':[1,0,0], 'b':[0,1,0], 'c':[1,0,0], 'd':[2,3,4]})
In [163]: df
Out[163]:
a b c d
0 1 0 1 2
1 0 1 0 3
2 0 0 0 4
In [164]: df.groupby(df.sum(), axis=1).sum()
Out[164]:
1 9
0 2 2
1 1 3
2 0 4
Returning a Series to propagate names¶
Group DataFrame columns, compute a set of metrics and return a named Series. The Series name is used as the name for the column index. This is especially useful in conjunction with reshaping operations such as stacking in which the column index name will be used as the name of the inserted column:
In [165]: df = pd.DataFrame({
.....: 'a': [0, 0, 0, 0, 1, 1, 1, 1, 2, 2, 2, 2],
.....: 'b': [0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 1, 1],
.....: 'c': [1, 0, 1, 0, 1, 0, 1, 0, 1, 0, 1, 0],
.....: 'd': [0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1],
.....: })
.....:
In [166]: def compute_metrics(x):
.....: result = {'b_sum': x['b'].sum(), 'c_mean': x['c'].mean()}
.....: return pd.Series(result, name='metrics')
.....:
In [167]: result = df.groupby('a').apply(compute_metrics)
In [168]: result
Out[168]:
metrics b_sum c_mean
a
0 2 0.5
1 2 0.5
2 2 0.5
In [169]: result.stack()
Out[169]:
a metrics
0 b_sum 2.0
c_mean 0.5
1 b_sum 2.0
c_mean 0.5
2 b_sum 2.0
c_mean 0.5
dtype: float64