Contributing to pandas¶
Table of contents:
Where to start?¶
All contributions, bug reports, bug fixes, documentation improvements, enhancements and ideas are welcome.
If you are simply looking to start working with the pandas codebase, navigate to the GitHub “issues” tab and start looking through interesting issues. There are a number of issues listed under Docs and Difficulty Novice where you could start out.
Or maybe through using pandas you have an idea of your own or are looking for something in the documentation and thinking ‘this can be improved’...you can do something about it!
Feel free to ask questions on the mailing list or on Gitter.
Bug reports and enhancement requests¶
Bug reports are an important part of making pandas more stable. Having a complete bug report will allow others to reproduce the bug and provide insight into fixing. Because many versions of pandas are supported, knowing version information will also identify improvements made since previous versions. Trying the bug-producing code out on the master branch is often a worthwhile exercise to confirm the bug still exists. It is also worth searching existing bug reports and pull requests to see if the issue has already been reported and/or fixed.
Bug reports must:
Include a short, self-contained Python snippet reproducing the problem. You can format the code nicely by using GitHub Flavored Markdown:
```python >>> from pandas import DataFrame >>> df = DataFrame(...) ... ```
Include the full version string of pandas and its dependencies. You can use the built in function:
>>> import pandas as pd >>> pd.show_versions()
Explain why the current behavior is wrong/not desired and what you expect instead.
The issue will then show up to the pandas community and be open to comments/ideas from others.
Working with the code¶
Now that you have an issue you want to fix, enhancement to add, or documentation to improve, you need to learn how to work with GitHub and the pandas code base.
Version control, Git, and GitHub¶
To the new user, working with Git is one of the more daunting aspects of contributing to pandas. It can very quickly become overwhelming, but sticking to the guidelines below will help keep the process straightforward and mostly trouble free. As always, if you are having difficulties please feel free to ask for help.
The code is hosted on GitHub. To contribute you will need to sign up for a free GitHub account. We use Git for version control to allow many people to work together on the project.
Some great resources for learning Git:
- the GitHub help pages.
- the NumPy’s documentation.
- Matthew Brett’s Pydagogue.
Getting started with Git¶
GitHub has instructions for installing git, setting up your SSH key, and configuring git. All these steps need to be completed before you can work seamlessly between your local repository and GitHub.
Forking¶
You will need your own fork to work on the code. Go to the pandas project
page and hit the Fork button. You will
want to clone your fork to your machine:
git clone https://github.com/your-user-name/pandas.git pandas-yourname
cd pandas-yourname
git remote add upstream git://github.com/pandas-dev/pandas.git
This creates the directory pandas-yourname and connects your repository to the upstream (main project) pandas repository.
Creating a branch¶
You want your master branch to reflect only production-ready code, so create a feature branch for making your changes. For example:
git branch shiny-new-feature
git checkout shiny-new-feature
The above can be simplified to:
git checkout -b shiny-new-feature
This changes your working directory to the shiny-new-feature branch. Keep any changes in this branch specific to one bug or feature so it is clear what the branch brings to pandas. You can have many shiny-new-features and switch in between them using the git checkout command.
To update this branch, you need to retrieve the changes from the master branch:
git fetch upstream
git rebase upstream/master
This will replay your commits on top of the latest pandas git master. If this
leads to merge conflicts, you must resolve these before submitting your pull
request. If you have uncommitted changes, you will need to stash them prior
to updating. This will effectively store your changes and they can be reapplied
after updating.
Creating a development environment¶
An easy way to create a pandas development environment is as follows.
- Install either Anaconda or miniconda
- Make sure that you have cloned the repository
cdto the pandas source directory
Tell conda to create a new environment, named pandas_dev, or any other name you would like
for this environment, by running:
conda create -n pandas_dev --file ci/requirements_dev.txt
For a python 3 environment:
conda create -n pandas_dev python=3 --file ci/requirements_dev.txt
Warning
If you are on Windows, see here for a fully compliant Windows environment.
This will create the new environment, and not touch any of your existing environments, nor any existing python installation. It will install all of the basic dependencies of pandas, as well as the development and testing tools. If you would like to install other dependencies, you can install them as follows:
conda install -n pandas_dev -c pandas pytables scipy
To install all pandas dependencies you can do the following:
conda install -n pandas_dev -c conda-forge --file ci/requirements_all.txt
To work in this environment, Windows users should activate it as follows:
activate pandas_dev
Mac OSX / Linux users should use:
source activate pandas_dev
You will then see a confirmation message to indicate you are in the new development environment.
To view your environments:
conda info -e
To return to your home root environment in Windows:
deactivate
To return to your home root environment in OSX / Linux:
source deactivate
See the full conda docs here.
At this point you can easily do an in-place install, as detailed in the next section.
Creating a Windows development environment¶
To build on Windows, you need to have compilers installed to build the extensions. You will need to install the appropriate Visual Studio compilers, VS 2008 for Python 2.7, VS 2010 for 3.4, and VS 2015 for Python 3.5 and 3.6.
For Python 2.7, you can install the mingw compiler which will work equivalently to VS 2008:
conda install -n pandas_dev libpython
or use the Microsoft Visual Studio VC++ compiler for Python. Note that you have to check the x64 box to install the x64 extension building capability as this is not installed by default.
For Python 3.4, you can download and install the Windows 7.1 SDK. Read the references below as there may be various gotchas during the installation.
For Python 3.5 and 3.6, you can download and install the Visual Studio 2015 Community Edition.
Here are some references and blogs:
- https://blogs.msdn.microsoft.com/pythonengineering/2016/04/11/unable-to-find-vcvarsall-bat/
- https://github.com/conda/conda-recipes/wiki/Building-from-Source-on-Windows-32-bit-and-64-bit
- https://cowboyprogrammer.org/building-python-wheels-for-windows/
- https://blog.ionelmc.ro/2014/12/21/compiling-python-extensions-on-windows/
- https://support.enthought.com/hc/en-us/articles/204469260-Building-Python-extensions-with-Canopy
Making changes¶
Before making your code changes, it is often necessary to build the code that was just checked out. There are two primary methods of doing this.
The best way to develop pandas is to build the C extensions in-place by running:
python setup.py build_ext --inplace
If you startup the Python interpreter in the pandas source directory you will call the built C extensions
Another very common option is to do a
developinstall of pandas:python setup.py develop
This makes a symbolic link that tells the Python interpreter to import pandas from your development directory. Thus, you can always be using the development version on your system without being inside the clone directory.
Contributing to the documentation¶
If you’re not the developer type, contributing to the documentation is still of huge value. You don’t even have to be an expert on pandas to do so! Something as simple as rewriting small passages for clarity as you reference the docs is a simple but effective way to contribute. The next person to read that passage will be in your debt!
In fact, there are sections of the docs that are worse off after being written by experts. If something in the docs doesn’t make sense to you, updating the relevant section after you figure it out is a simple way to ensure it will help the next person.
Documentation:
About the pandas documentation¶
The documentation is written in reStructuredText, which is almost like writing in plain English, and built using Sphinx. The Sphinx Documentation has an excellent introduction to reST. Review the Sphinx docs to perform more complex changes to the documentation as well.
Some other important things to know about the docs:
The pandas documentation consists of two parts: the docstrings in the code itself and the docs in this folder
pandas/doc/.The docstrings provide a clear explanation of the usage of the individual functions, while the documentation in this folder consists of tutorial-like overviews per topic together with some other information (what’s new, installation, etc).
The docstrings follow the Numpy Docstring Standard, which is used widely in the Scientific Python community. This standard specifies the format of the different sections of the docstring. See this document for a detailed explanation, or look at some of the existing functions to extend it in a similar manner.
The tutorials make heavy use of the ipython directive sphinx extension. This directive lets you put code in the documentation which will be run during the doc build. For example:
.. ipython:: python x = 2 x**3
will be rendered as:
In [1]: x = 2 In [2]: x**3 Out[2]: 8
Almost all code examples in the docs are run (and the output saved) during the doc build. This approach means that code examples will always be up to date, but it does make the doc building a bit more complex.
Note
The .rst files are used to automatically generate Markdown and HTML versions
of the docs. For this reason, please do not edit CONTRIBUTING.md directly,
but instead make any changes to doc/source/contributing.rst. Then, to
generate CONTRIBUTING.md, use pandoc
with the following command:
pandoc doc/source/contributing.rst -t markdown_github > CONTRIBUTING.md
The utility script scripts/api_rst_coverage.py can be used to compare
the list of methods documented in doc/source/api.rst (which is used to generate
the API Reference page)
and the actual public methods.
This will identify methods documented in in doc/source/api.rst that are not actually
class methods, and existing methods that are not documented in doc/source/api.rst.
How to build the pandas documentation¶
Requirements¶
First, you need to have a development environment to be able to build pandas
(see the docs on creating a development environment above).
Further, to build the docs, there are some extra requirements: you will need to
have sphinx and ipython installed. numpydoc is used to parse the docstrings that
follow the Numpy Docstring Standard (see above), but you don’t need to install
this because a local copy of numpydoc is included in the pandas source
code. nbsphinx is required to build
the Jupyter notebooks included in the documentation.
If you have a conda environment named pandas_dev, you can install the extra
requirements with:
conda install -n pandas_dev sphinx ipython nbconvert nbformat
conda install -n pandas_dev -c conda-forge nbsphinx
Furthermore, it is recommended to have all optional dependencies.
installed. This is not strictly necessary, but be aware that you will see some error
messages when building the docs. This happens because all the code in the documentation
is executed during the doc build, and so code examples using optional dependencies
will generate errors. Run pd.show_versions() to get an overview of the installed
version of all dependencies.
Warning
You need to have sphinx version >= 1.3.2.
Building the documentation¶
So how do you build the docs? Navigate to your local
pandas/doc/ directory in the console and run:
python make.py html
Then you can find the HTML output in the folder pandas/doc/build/html/.
The first time you build the docs, it will take quite a while because it has to run all the code examples and build all the generated docstring pages. In subsequent evocations, sphinx will try to only build the pages that have been modified.
If you want to do a full clean build, do:
python make.py clean
python make.py html
Starting with pandas 0.13.1 you can tell make.py to compile only a single section
of the docs, greatly reducing the turn-around time for checking your changes.
You will be prompted to delete .rst files that aren’t required. This is okay because
the prior versions of these files can be checked out from git. However, you must make sure
not to commit the file deletions to your Git repository!
#omit autosummary and API section
python make.py clean
python make.py --no-api
# compile the docs with only a single
# section, that which is in indexing.rst
python make.py clean
python make.py --single indexing
For comparison, a full documentation build may take 10 minutes, a -no-api build
may take 3 minutes and a single section may take 15 seconds. Subsequent builds, which
only process portions you have changed, will be faster. Open the following file in a web
browser to see the full documentation you just built:
pandas/docs/build/html/index.html
And you’ll have the satisfaction of seeing your new and improved documentation!
Building master branch documentation¶
When pull requests are merged into the pandas master branch, the main parts of
the documentation are also built by Travis-CI. These docs are then hosted here, see also
the Continuous Integration section.
Contributing to the code base¶
Code Base:
Code standards¶
Writing good code is not just about what you write. It is also about how you write it. During Continuous Integration testing, several tools will be run to check your code for stylistic errors. Generating any warnings will cause the test to fail. Thus, good style is a requirement for submitting code to pandas.
In addition, because a lot of people use our library, it is important that we do not make sudden changes to the code that could have the potential to break a lot of user code as a result, that is, we need it to be as backwards compatible as possible to avoid mass breakages.
Additional standards are outlined on the code style wiki page.
C (cpplint)¶
pandas uses the Google
standard. Google provides an open source style checker called cpplint, but we
use a fork of it that can be found here.
Here are some of the more common cpplint issues:
- we restrict line-length to 80 characters to promote readability
- every header file must include a header guard to avoid name collisions if re-included
Continuous Integration. will run the cpplint tool and report any stylistic errors in your code. Therefore, it is helpful before submitting code to run the check yourself:
cpplint --extensions=c,h --headers=h --filter=-readability/casting,-runtime/int,-build/include_subdir modified-c-file
You can also run this command on an entire directory if necessary:
cpplint --extensions=c,h --headers=h --filter=-readability/casting,-runtime/int,-build/include_subdir --recursive modified-c-directory
To make your commits compliant with this standard, you can install the ClangFormat tool, which can be downloaded here. To configure, in your home directory, run the following command:
clang-format style=google -dump-config > .clang-format
Then modify the file to ensure that any indentation width parameters are at least four. Once configured, you can run the tool as follows:
clang-format modified-c-file
This will output what your file will look like if the changes are made, and to apply them, just run the following command:
clang-format -i modified-c-file
To run the tool on an entire directory, you can run the following analogous commands:
clang-format modified-c-directory/*.c modified-c-directory/*.h
clang-format -i modified-c-directory/*.c modified-c-directory/*.h
Do note that this tool is best-effort, meaning that it will try to correct as
many errors as possible, but it may not correct all of them. Thus, it is
recommended that you run cpplint to double check and make any other style
fixes manually.
Python (PEP8)¶
pandas uses the PEP8 standard.
There are several tools to ensure you abide by this standard. Here are some of
the more common PEP8 issues:
- we restrict line-length to 79 characters to promote readability
- passing arguments should have spaces after commas, e.g.
foo(arg1, arg2, kw1='bar')
Continuous Integration will run the flake8 tool and report any stylistic errors in your code. Therefore, it is helpful before submitting code to run the check yourself on the diff:
git diff master -u -- "*.py" | flake8 --diff
This command will catch any stylistic errors in your changes specifically, but be beware it may not catch all of them. For example, if you delete the only usage of an imported function, it is stylistically incorrect to import an unused function. However, style-checking the diff will not catch this because the actual import is not part of the diff. Thus, for completeness, you should run this command, though it will take longer:
git diff master --name-only -- "*.py" | grep "pandas/" | xargs -r flake8
Note that on OSX, the -r flag is not available, so you have to omit it and
run this slightly modified command:
git diff master --name-only -- "*.py" | grep "pandas/" | xargs flake8
Note that on Windows, these commands are unfortunately not possible because
commands like grep and xargs are not available natively. To imitate the
behavior with the commands above, you should run:
git diff master --name-only -- "*.py"
This will list all of the Python files that have been modified. The only ones
that matter during linting are any whose directory filepath begins with “pandas.”
For each filepath, copy and paste it after the flake8 command as shown below:
flake8 <python-filepath>
Alternatively, you can install the grep and xargs commands via the
MinGW toolchain, and it will allow you to run the
commands above.
Backwards Compatibility¶
Please try to maintain backward compatibility. pandas has lots of users with lots of existing code, so don’t break it if at all possible. If you think breakage is required, clearly state why as part of the pull request. Also, be careful when changing method signatures and add deprecation warnings where needed.
Testing With Continuous Integration¶
The pandas test suite will run automatically on Travis-CI, Appveyor, and Circle CI continuous integration services, once your pull request is submitted. However, if you wish to run the test suite on a branch prior to submitting the pull request, then the continuous integration services need to be hooked to your GitHub repository. Instructions are here for Travis-CI, Appveyor , and CircleCI.
A pull-request will be considered for merging when you have an all ‘green’ build. If any tests are failing, then you will get a red ‘X’, where you can click through to see the individual failed tests. This is an example of a green build.
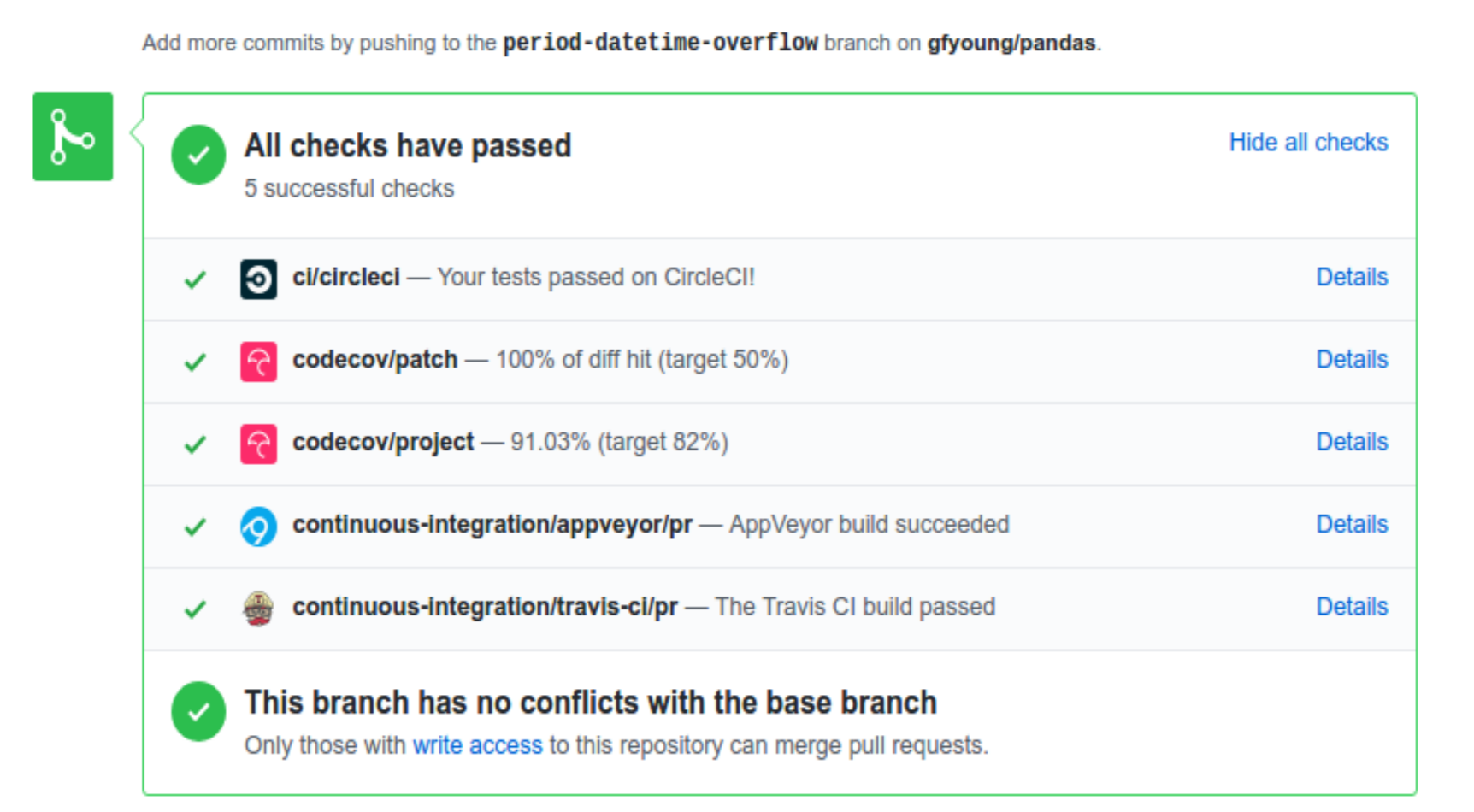
Note
Each time you push to your fork, a new run of the tests will be triggered on the CI. Appveyor will auto-cancel any non-currently-running tests for that same pull-request. You can enable the auto-cancel feature for Travis-CI here and for CircleCI here.
Test-driven development/code writing¶
pandas is serious about testing and strongly encourages contributors to embrace test-driven development (TDD). This development process “relies on the repetition of a very short development cycle: first the developer writes an (initially failing) automated test case that defines a desired improvement or new function, then produces the minimum amount of code to pass that test.” So, before actually writing any code, you should write your tests. Often the test can be taken from the original GitHub issue. However, it is always worth considering additional use cases and writing corresponding tests.
Adding tests is one of the most common requests after code is pushed to pandas. Therefore, it is worth getting in the habit of writing tests ahead of time so this is never an issue.
Like many packages, pandas uses pytest and the convenient extensions in numpy.testing.
Note
The earliest supported pytest version is 3.1.0.
Writing tests¶
All tests should go into the tests subdirectory of the specific package.
This folder contains many current examples of tests, and we suggest looking to these for
inspiration. If your test requires working with files or
network connectivity, there is more information on the testing page of the wiki.
The pandas.util.testing module has many special assert functions that
make it easier to make statements about whether Series or DataFrame objects are
equivalent. The easiest way to verify that your code is correct is to
explicitly construct the result you expect, then compare the actual result to
the expected correct result:
def test_pivot(self):
data = {
'index' : ['A', 'B', 'C', 'C', 'B', 'A'],
'columns' : ['One', 'One', 'One', 'Two', 'Two', 'Two'],
'values' : [1., 2., 3., 3., 2., 1.]
}
frame = DataFrame(data)
pivoted = frame.pivot(index='index', columns='columns', values='values')
expected = DataFrame({
'One' : {'A' : 1., 'B' : 2., 'C' : 3.},
'Two' : {'A' : 1., 'B' : 2., 'C' : 3.}
})
assert_frame_equal(pivoted, expected)
Transitioning to pytest¶
pandas existing test structure is mostly classed based, meaning that you will typically find tests wrapped in a class.
class TestReallyCoolFeature(object):
....
Going forward, we are moving to a more functional style using the pytest framework, which offers a richer testing framework that will facilitate testing and developing. Thus, instead of writing test classes, we will write test functions like this:
def test_really_cool_feature():
....
Using pytest¶
Here is an example of a self-contained set of tests that illustrate multiple features that we like to use.
- functional style: tests are like
test_*and only take arguments that are either fixtures or parameters pytest.markcan be used to set metadata on test functions, e.g.skiporxfail.- using
parametrize: allow testing of multiple cases - to set a mark on a parameter,
pytest.param(..., marks=...)syntax should be used fixture, code for object construction, on a per-test basis- using bare
assertfor scalars and truth-testing tm.assert_series_equal(and its counter parttm.assert_frame_equal), for pandas object comparisons.- the typical pattern of constructing an
expectedand comparing versus theresult
We would name this file test_cool_feature.py and put in an appropriate place in the pandas/tests/ structure.
import pytest
import numpy as np
import pandas as pd
from pandas.util import testing as tm
@pytest.mark.parametrize('dtype', ['int8', 'int16', 'int32', 'int64'])
def test_dtypes(dtype):
assert str(np.dtype(dtype)) == dtype
@pytest.mark.parametrize('dtype', ['float32',
pytest.param('int16', marks=pytest.mark.skip),
pytest.param('int32',
marks=pytest.mark.xfail(reason='to show how it works'))])
def test_mark(dtype):
assert str(np.dtype(dtype)) == 'float32'
@pytest.fixture
def series():
return pd.Series([1, 2, 3])
@pytest.fixture(params=['int8', 'int16', 'int32', 'int64'])
def dtype(request):
return request.param
def test_series(series, dtype):
result = series.astype(dtype)
assert result.dtype == dtype
expected = pd.Series([1, 2, 3], dtype=dtype)
tm.assert_series_equal(result, expected)
A test run of this yields
((pandas) bash-3.2$ pytest test_cool_feature.py -v
=========================== test session starts ===========================
platform darwin -- Python 3.6.2, pytest-3.2.1, py-1.4.31, pluggy-0.4.0
collected 11 items
tester.py::test_dtypes[int8] PASSED
tester.py::test_dtypes[int16] PASSED
tester.py::test_dtypes[int32] PASSED
tester.py::test_dtypes[int64] PASSED
tester.py::test_mark[float32] PASSED
tester.py::test_mark[int16] SKIPPED
tester.py::test_mark[int32] xfail
tester.py::test_series[int8] PASSED
tester.py::test_series[int16] PASSED
tester.py::test_series[int32] PASSED
tester.py::test_series[int64] PASSED
Tests that we have parametrized are now accessible via the test name, for example we could run these with -k int8 to sub-select only those tests which match int8.
((pandas) bash-3.2$ pytest test_cool_feature.py -v -k int8
=========================== test session starts ===========================
platform darwin -- Python 3.6.2, pytest-3.2.1, py-1.4.31, pluggy-0.4.0
collected 11 items
test_cool_feature.py::test_dtypes[int8] PASSED
test_cool_feature.py::test_series[int8] PASSED
Running the test suite¶
The tests can then be run directly inside your Git clone (without having to install pandas) by typing:
pytest pandas
The tests suite is exhaustive and takes around 20 minutes to run. Often it is worth running only a subset of tests first around your changes before running the entire suite.
The easiest way to do this is with:
pytest pandas/path/to/test.py -k regex_matching_test_name
Or with one of the following constructs:
pytest pandas/tests/[test-module].py
pytest pandas/tests/[test-module].py::[TestClass]
pytest pandas/tests/[test-module].py::[TestClass]::[test_method]
Using pytest-xdist, one can speed up local testing on multicore machines. To use this feature, you will need to install pytest-xdist via:
pip install pytest-xdist
Two scripts are provided to assist with this. These scripts distribute testing across 4 threads.
On Unix variants, one can type:
test_fast.sh
On Windows, one can type:
test_fast.bat
This can significantly reduce the time it takes to locally run tests before submitting a pull request.
For more, see the pytest documentation.
New in version 0.20.0.
Furthermore one can run
pd.test()
with an imported pandas to run tests similarly.
Running the performance test suite¶
Performance matters and it is worth considering whether your code has introduced
performance regressions. pandas is in the process of migrating to
asv benchmarks
to enable easy monitoring of the performance of critical pandas operations.
These benchmarks are all found in the pandas/asv_bench directory. asv
supports both python2 and python3.
To use all features of asv, you will need either conda or
virtualenv. For more details please check the asv installation
webpage.
To install asv:
pip install git+https://github.com/spacetelescope/asv
If you need to run a benchmark, change your directory to asv_bench/ and run:
asv continuous -f 1.1 upstream/master HEAD
You can replace HEAD with the name of the branch you are working on,
and report benchmarks that changed by more than 10%.
The command uses conda by default for creating the benchmark
environments. If you want to use virtualenv instead, write:
asv continuous -f 1.1 -E virtualenv upstream/master HEAD
The -E virtualenv option should be added to all asv commands
that run benchmarks. The default value is defined in asv.conf.json.
Running the full test suite can take up to one hour and use up to 3GB of RAM.
Usually it is sufficient to paste only a subset of the results into the pull
request to show that the committed changes do not cause unexpected performance
regressions. You can run specific benchmarks using the -b flag, which
takes a regular expression. For example, this will only run tests from a
pandas/asv_bench/benchmarks/groupby.py file:
asv continuous -f 1.1 upstream/master HEAD -b ^groupby
If you want to only run a specific group of tests from a file, you can do it
using . as a separator. For example:
asv continuous -f 1.1 upstream/master HEAD -b groupby.groupby_agg_builtins
will only run the groupby_agg_builtins benchmark defined in groupby.py.
You can also run the benchmark suite using the version of pandas
already installed in your current Python environment. This can be
useful if you do not have virtualenv or conda, or are using the
setup.py develop approach discussed above; for the in-place build
you need to set PYTHONPATH, e.g.
PYTHONPATH="$PWD/.." asv [remaining arguments].
You can run benchmarks using an existing Python
environment by:
asv run -e -E existing
or, to use a specific Python interpreter,:
asv run -e -E existing:python3.5
This will display stderr from the benchmarks, and use your local
python that comes from your $PATH.
Information on how to write a benchmark and how to use asv can be found in the asv documentation.
Documenting your code¶
Changes should be reflected in the release notes located in doc/source/whatsnew/vx.y.z.txt.
This file contains an ongoing change log for each release. Add an entry to this file to
document your fix, enhancement or (unavoidable) breaking change. Make sure to include the
GitHub issue number when adding your entry (using `` GH1234 `` where 1234 is the
issue/pull request number).
If your code is an enhancement, it is most likely necessary to add usage
examples to the existing documentation. This can be done following the section
regarding documentation above.
Further, to let users know when this feature was added, the versionadded
directive is used. The sphinx syntax for that is:
.. versionadded:: 0.17.0
This will put the text New in version 0.17.0 wherever you put the sphinx directive. This should also be put in the docstring when adding a new function or method (example) or a new keyword argument (example).
Contributing your changes to pandas¶
Committing your code¶
Keep style fixes to a separate commit to make your pull request more readable.
Once you’ve made changes, you can see them by typing:
git status
If you have created a new file, it is not being tracked by git. Add it by typing:
git add path/to/file-to-be-added.py
Doing ‘git status’ again should give something like:
# On branch shiny-new-feature
#
# modified: /relative/path/to/file-you-added.py
#
Finally, commit your changes to your local repository with an explanatory message. Pandas uses a convention for commit message prefixes and layout. Here are some common prefixes along with general guidelines for when to use them:
- ENH: Enhancement, new functionality
- BUG: Bug fix
- DOC: Additions/updates to documentation
- TST: Additions/updates to tests
- BLD: Updates to the build process/scripts
- PERF: Performance improvement
- CLN: Code cleanup
The following defines how a commit message should be structured. Please reference the relevant GitHub issues in your commit message using GH1234 or #1234. Either style is fine, but the former is generally preferred:
- a subject line with < 80 chars.
- One blank line.
- Optionally, a commit message body.
Now you can commit your changes in your local repository:
git commit -m
Combining commits¶
If you have multiple commits, you may want to combine them into one commit, often referred to as “squashing” or “rebasing”. This is a common request by package maintainers when submitting a pull request as it maintains a more compact commit history. To rebase your commits:
git rebase -i HEAD~#
Where # is the number of commits you want to combine. Then you can pick the relevant commit message and discard others.
To squash to the master branch do:
git rebase -i master
Use the s option on a commit to squash, meaning to keep the commit messages,
or f to fixup, meaning to merge the commit messages.
Then you will need to push the branch (see below) forcefully to replace the current commits with the new ones:
git push origin shiny-new-feature -f
Pushing your changes¶
When you want your changes to appear publicly on your GitHub page, push your forked feature branch’s commits:
git push origin shiny-new-feature
Here origin is the default name given to your remote repository on GitHub.
You can see the remote repositories:
git remote -v
If you added the upstream repository as described above you will see something like:
origin git@github.com:yourname/pandas.git (fetch)
origin git@github.com:yourname/pandas.git (push)
upstream git://github.com/pandas-dev/pandas.git (fetch)
upstream git://github.com/pandas-dev/pandas.git (push)
Now your code is on GitHub, but it is not yet a part of the pandas project. For that to happen, a pull request needs to be submitted on GitHub.
Review your code¶
When you’re ready to ask for a code review, file a pull request. Before you do, once again make sure that you have followed all the guidelines outlined in this document regarding code style, tests, performance tests, and documentation. You should also double check your branch changes against the branch it was based on:
- Navigate to your repository on GitHub – https://github.com/your-user-name/pandas
- Click on
Branches - Click on the
Comparebutton for your feature branch - Select the
baseandcomparebranches, if necessary. This will bemasterandshiny-new-feature, respectively.
Finally, make the pull request¶
If everything looks good, you are ready to make a pull request. A pull request is how code from a local repository becomes available to the GitHub community and can be looked at and eventually merged into the master version. This pull request and its associated changes will eventually be committed to the master branch and available in the next release. To submit a pull request:
- Navigate to your repository on GitHub
- Click on the
Pull Requestbutton - You can then click on
CommitsandFiles Changedto make sure everything looks okay one last time - Write a description of your changes in the
Preview Discussiontab - Click
Send Pull Request.
This request then goes to the repository maintainers, and they will review the code. If you need to make more changes, you can make them in your branch, push them to GitHub, and the pull request will be automatically updated. Pushing them to GitHub again is done by:
git push -f origin shiny-new-feature
This will automatically update your pull request with the latest code and restart the Continuous Integration tests.
Delete your merged branch (optional)¶
Once your feature branch is accepted into upstream, you’ll probably want to get rid of the branch. First, merge upstream master into your branch so git knows it is safe to delete your branch:
git fetch upstream
git checkout master
git merge upstream/master
Then you can just do:
git branch -d shiny-new-feature
Make sure you use a lower-case -d, or else git won’t warn you if your feature
branch has not actually been merged.
The branch will still exist on GitHub, so to delete it there do:
git push origin --delete shiny-new-feature