What’s New¶
These are new features and improvements of note in each release.
v0.11.0 (April 22, 2013)¶
This is a major release from 0.10.1 and includes many new features and enhancements along with a large number of bug fixes. The methods of Selecting Data have had quite a number of additions, and Dtype support is now full-fledged. There are also a number of important API changes that long-time pandas users should pay close attention to.
There is a new section in the documentation, 10 Minutes to Pandas, primarily geared to new users.
There is a new section in the documentation, Cookbook, a collection of useful recipes in pandas (and that we want contributions!).
There are several libraries that are now Recommended Dependencies
Selection Choices¶
Starting in 0.11.0, object selection has had a number of user-requested additions in order to support more explicit location based indexing. Pandas now supports three types of multi-axis indexing.
.loc is strictly label based, will raise KeyError when the items are not found, allowed inputs are:
- A single label, e.g. 5 or 'a', (note that 5 is interpreted as a label of the index. This use is not an integer position along the index)
- A list or array of labels ['a', 'b', 'c']
- A slice object with labels 'a':'f', (note that contrary to usual python slices, both the start and the stop are included!)
- A boolean array
See more at Selection by Label
.iloc is strictly integer position based (from 0 to length-1 of the axis), will raise IndexError when the requested indicies are out of bounds. Allowed inputs are:
- An integer e.g. 5
- A list or array of integers [4, 3, 0]
- A slice object with ints 1:7
- A boolean array
See more at Selection by Position
.ix supports mixed integer and label based access. It is primarily label based, but will fallback to integer positional access. .ix is the most general and will support any of the inputs to .loc and .iloc, as well as support for floating point label schemes. .ix is especially useful when dealing with mixed positional and label based hierarchial indexes.
As using integer slices with .ix have different behavior depending on whether the slice is interpreted as position based or label based, it’s usually better to be explicit and use .iloc or .loc.
See more at Advanced Indexing, Advanced Hierarchical and Fallback Indexing
Selection Deprecations¶
Starting in version 0.11.0, these methods may be deprecated in future versions.
- irow
- icol
- iget_value
See the section Selection by Position for substitutes.
Dtypes¶
Numeric dtypes will propagate and can coexist in DataFrames. If a dtype is passed (either directly via the dtype keyword, a passed ndarray, or a passed Series, then it will be preserved in DataFrame operations. Furthermore, different numeric dtypes will NOT be combined. The following example will give you a taste.
In [1808]: df1 = DataFrame(randn(8, 1), columns = ['A'], dtype = 'float32')
In [1809]: df1
Out[1809]:
A
0 0.741687
1 0.035967
2 -2.700230
3 0.777316
4 1.201654
5 0.775594
6 0.916695
7 -0.511978
In [1810]: df1.dtypes
Out[1810]:
A float32
dtype: object
In [1811]: df2 = DataFrame(dict( A = Series(randn(8),dtype='float16'),
......: B = Series(randn(8)),
......: C = Series(randn(8),dtype='uint8') ))
......:
In [1812]: df2
Out[1812]:
A B C
0 0.805664 -1.750153 0
1 -0.517578 0.507924 0
2 -0.980469 -0.163195 0
3 -1.325195 0.285564 255
4 0.015396 -0.332279 0
5 1.063477 -0.516040 0
6 -0.297363 -0.531297 0
7 1.118164 -0.409554 0
In [1813]: df2.dtypes
Out[1813]:
A float16
B float64
C uint8
dtype: object
# here you get some upcasting
In [1814]: df3 = df1.reindex_like(df2).fillna(value=0.0) + df2
In [1815]: df3
Out[1815]:
A B C
0 1.547351 -1.750153 0
1 -0.481611 0.507924 0
2 -3.680699 -0.163195 0
3 -0.547880 0.285564 255
4 1.217050 -0.332279 0
5 1.839071 -0.516040 0
6 0.619332 -0.531297 0
7 0.606186 -0.409554 0
In [1816]: df3.dtypes
Out[1816]:
A float32
B float64
C float64
dtype: object
Dtype Conversion¶
This is lower-common-denomicator upcasting, meaning you get the dtype which can accomodate all of the types
In [1817]: df3.values.dtype
Out[1817]: dtype('float64')
Conversion
In [1818]: df3.astype('float32').dtypes
Out[1818]:
A float32
B float32
C float32
dtype: object
Mixed Conversion
In [1819]: df3['D'] = '1.'
In [1820]: df3['E'] = '1'
In [1821]: df3.convert_objects(convert_numeric=True).dtypes
Out[1821]:
A float32
B float64
C float64
D float64
E int64
dtype: object
# same, but specific dtype conversion
In [1822]: df3['D'] = df3['D'].astype('float16')
In [1823]: df3['E'] = df3['E'].astype('int32')
In [1824]: df3.dtypes
Out[1824]:
A float32
B float64
C float64
D float16
E int32
dtype: object
Forcing Date coercion (and setting NaT when not datelike)
In [1825]: s = Series([datetime(2001,1,1,0,0), 'foo', 1.0, 1,
......: Timestamp('20010104'), '20010105'],dtype='O')
......:
In [1826]: s.convert_objects(convert_dates='coerce')
Out[1826]:
0 2001-01-01 00:00:00
1 NaT
2 NaT
3 NaT
4 2001-01-04 00:00:00
5 2001-01-05 00:00:00
dtype: datetime64[ns]
Dtype Gotchas¶
Platform Gotchas
Starting in 0.11.0, construction of DataFrame/Series will use default dtypes of int64 and float64, regardless of platform. This is not an apparent change from earlier versions of pandas. If you specify dtypes, they WILL be respected, however (GH2837)
The following will all result in int64 dtypes
In [1827]: DataFrame([1,2],columns=['a']).dtypes
Out[1827]:
a int64
dtype: object
In [1828]: DataFrame({'a' : [1,2] }).dtypes
Out[1828]:
a int64
dtype: object
In [1829]: DataFrame({'a' : 1 }, index=range(2)).dtypes
Out[1829]:
a int64
dtype: object
Keep in mind that DataFrame(np.array([1,2])) WILL result in int32 on 32-bit platforms!
Upcasting Gotchas
Performing indexing operations on integer type data can easily upcast the data. The dtype of the input data will be preserved in cases where nans are not introduced.
In [1830]: dfi = df3.astype('int32')
In [1831]: dfi['D'] = dfi['D'].astype('int64')
In [1832]: dfi
Out[1832]:
A B C D E
0 1 -1 0 1 1
1 0 0 0 1 1
2 -3 0 0 1 1
3 0 0 255 1 1
4 1 0 0 1 1
5 1 0 0 1 1
6 0 0 0 1 1
7 0 0 0 1 1
In [1833]: dfi.dtypes
Out[1833]:
A int32
B int32
C int32
D int64
E int32
dtype: object
In [1834]: casted = dfi[dfi>0]
In [1835]: casted
Out[1835]:
A B C D E
0 1 NaN NaN 1 1
1 NaN NaN NaN 1 1
2 NaN NaN NaN 1 1
3 NaN NaN 255 1 1
4 1 NaN NaN 1 1
5 1 NaN NaN 1 1
6 NaN NaN NaN 1 1
7 NaN NaN NaN 1 1
In [1836]: casted.dtypes
Out[1836]:
A float64
B float64
C float64
D int64
E int32
dtype: object
While float dtypes are unchanged.
In [1837]: df4 = df3.copy()
In [1838]: df4['A'] = df4['A'].astype('float32')
In [1839]: df4.dtypes
Out[1839]:
A float32
B float64
C float64
D float16
E int32
dtype: object
In [1840]: casted = df4[df4>0]
In [1841]: casted
Out[1841]:
A B C D E
0 1.547351 NaN NaN 1 1
1 NaN 0.507924 NaN 1 1
2 NaN NaN NaN 1 1
3 NaN 0.285564 255 1 1
4 1.217050 NaN NaN 1 1
5 1.839071 NaN NaN 1 1
6 0.619332 NaN NaN 1 1
7 0.606186 NaN NaN 1 1
In [1842]: casted.dtypes
Out[1842]:
A float32
B float64
C float64
D float16
E int32
dtype: object
Datetimes Conversion¶
Datetime64[ns] columns in a DataFrame (or a Series) allow the use of np.nan to indicate a nan value, in addition to the traditional NaT, or not-a-time. This allows convenient nan setting in a generic way. Furthermore datetime64[ns] columns are created by default, when passed datetimelike objects (this change was introduced in 0.10.1) (GH2809, GH2810)
In [1843]: df = DataFrame(randn(6,2),date_range('20010102',periods=6),columns=['A','B'])
In [1844]: df['timestamp'] = Timestamp('20010103')
In [1845]: df
Out[1845]:
A B timestamp
2001-01-02 0.175289 -0.961203 2001-01-03 00:00:00
2001-01-03 -0.302857 0.047525 2001-01-03 00:00:00
2001-01-04 -0.987381 -0.082381 2001-01-03 00:00:00
2001-01-05 1.122844 0.357760 2001-01-03 00:00:00
2001-01-06 -1.287685 -0.555503 2001-01-03 00:00:00
2001-01-07 -1.721204 -0.040879 2001-01-03 00:00:00
# datetime64[ns] out of the box
In [1846]: df.get_dtype_counts()
Out[1846]:
datetime64[ns] 1
float64 2
dtype: int64
# use the traditional nan, which is mapped to NaT internally
In [1847]: df.ix[2:4,['A','timestamp']] = np.nan
In [1848]: df
Out[1848]:
A B timestamp
2001-01-02 0.175289 -0.961203 2001-01-03 00:00:00
2001-01-03 -0.302857 0.047525 2001-01-03 00:00:00
2001-01-04 NaN -0.082381 NaT
2001-01-05 NaN 0.357760 NaT
2001-01-06 -1.287685 -0.555503 2001-01-03 00:00:00
2001-01-07 -1.721204 -0.040879 2001-01-03 00:00:00
Astype conversion on datetime64[ns] to object, implicity converts NaT to np.nan
In [1849]: import datetime
In [1850]: s = Series([datetime.datetime(2001, 1, 2, 0, 0) for i in range(3)])
In [1851]: s.dtype
Out[1851]: dtype('<M8[ns]')
In [1852]: s[1] = np.nan
In [1853]: s
Out[1853]:
0 2001-01-02 00:00:00
1 NaT
2 2001-01-02 00:00:00
dtype: datetime64[ns]
In [1854]: s.dtype
Out[1854]: dtype('<M8[ns]')
In [1855]: s = s.astype('O')
In [1856]: s
Out[1856]:
0 2001-01-02 00:00:00
1 NaN
2 2001-01-02 00:00:00
dtype: object
In [1857]: s.dtype
Out[1857]: dtype('O')
API changes¶
- Added to_series() method to indicies, to facilitate the creation of indexers (GH3275)
- HDFStore
- added the method select_column to select a single column from a table as a Series.
- deprecated the unique method, can be replicated by select_column(key,column).unique()
- min_itemsize parameter to append will now automatically create data_columns for passed keys
Enhancements¶
Improved performance of df.to_csv() by up to 10x in some cases. (GH3059)
Numexpr is now a Recommended Dependencies, to accelerate certain types of numerical and boolean operations
Bottleneck is now a Recommended Dependencies, to accelerate certain types of nan operations
HDFStore
support read_hdf/to_hdf API similar to read_csv/to_csv
In [1858]: df = DataFrame(dict(A=range(5), B=range(5))) In [1859]: df.to_hdf('store.h5','table',append=True) In [1860]: read_hdf('store.h5', 'table', where = ['index>2']) Out[1860]: A B 3 3 3 4 4 4provide dotted attribute access to get from stores, e.g. store.df == store['df']
new keywords iterator=boolean, and chunksize=number_in_a_chunk are provided to support iteration on select and select_as_multiple (GH3076)
You can now select timestamps from an unordered timeseries similarly to an ordered timeseries (GH2437)
You can now select with a string from a DataFrame with a datelike index, in a similar way to a Series (GH3070)
In [1861]: idx = date_range("2001-10-1", periods=5, freq='M') In [1862]: ts = Series(np.random.rand(len(idx)),index=idx) In [1863]: ts['2001'] Out[1863]: 2001-10-31 0.407874 2001-11-30 0.372920 2001-12-31 0.714280 Freq: M, dtype: float64 In [1864]: df = DataFrame(dict(A = ts)) In [1865]: df['2001'] Out[1865]: A 2001-10-31 0.407874 2001-11-30 0.372920 2001-12-31 0.714280Squeeze to possibly remove length 1 dimensions from an object.
In [1866]: p = Panel(randn(3,4,4),items=['ItemA','ItemB','ItemC'], ......: major_axis=date_range('20010102',periods=4), ......: minor_axis=['A','B','C','D']) ......: In [1867]: p Out[1867]: <class 'pandas.core.panel.Panel'> Dimensions: 3 (items) x 4 (major_axis) x 4 (minor_axis) Items axis: ItemA to ItemC Major_axis axis: 2001-01-02 00:00:00 to 2001-01-05 00:00:00 Minor_axis axis: A to D In [1868]: p.reindex(items=['ItemA']).squeeze() Out[1868]: A B C D 2001-01-02 1.799989 -1.604955 -0.300943 -0.037085 2001-01-03 1.153518 -1.207366 1.061454 0.713368 2001-01-04 -0.207985 1.232183 0.448277 1.277114 2001-01-05 0.089381 -1.350877 -1.529130 -1.007310 In [1869]: p.reindex(items=['ItemA'],minor=['B']).squeeze() Out[1869]: 2001-01-02 -1.604955 2001-01-03 -1.207366 2001-01-04 1.232183 2001-01-05 -1.350877 Freq: D, Name: B, dtype: float64In pd.io.data.Options,
- Fix bug when trying to fetch data for the current month when already past expiry.
- Now using lxml to scrape html instead of BeautifulSoup (lxml was faster).
- New instance variables for calls and puts are automatically created when a method that creates them is called. This works for current month where the instance variables are simply calls and puts. Also works for future expiry months and save the instance variable as callsMMYY or putsMMYY, where MMYY are, respectively, the month and year of the option’s expiry.
- Options.get_near_stock_price now allows the user to specify the month for which to get relevant options data.
- Options.get_forward_data now has optional kwargs near and above_below. This allows the user to specify if they would like to only return forward looking data for options near the current stock price. This just obtains the data from Options.get_near_stock_price instead of Options.get_xxx_data() (GH2758).
Cursor coordinate information is now displayed in time-series plots.
added option display.max_seq_items to control the number of elements printed per sequence pprinting it. (GH2979)
added option display.chop_threshold to control display of small numerical values. (GH2739)
added option display.max_info_rows to prevent verbose_info from being calculated for frames above 1M rows (configurable). (GH2807, GH2918)
value_counts() now accepts a “normalize” argument, for normalized histograms. (GH2710).
DataFrame.from_records now accepts not only dicts but any instance of the collections.Mapping ABC.
added option display.with_wmp_style providing a sleeker visual style for plots. Based on https://gist.github.com/huyng/816622 (GH3075).
Treat boolean values as integers (values 1 and 0) for numeric operations. (GH2641)
to_html() now accepts an optional “escape” argument to control reserved HTML character escaping (enabled by default) and escapes &, in addition to < and >. (GH2919)
See the full release notes or issue tracker on GitHub for a complete list.
v0.10.1 (January 22, 2013)¶
This is a minor release from 0.10.0 and includes new features, enhancements, and bug fixes. In particular, there is substantial new HDFStore functionality contributed by Jeff Reback.
An undesired API breakage with functions taking the inplace option has been reverted and deprecation warnings added.
API changes¶
- Functions taking an inplace option return the calling object as before. A deprecation message has been added
- Groupby aggregations Max/Min no longer exclude non-numeric data (GH2700)
- Resampling an empty DataFrame now returns an empty DataFrame instead of raising an exception (GH2640)
- The file reader will now raise an exception when NA values are found in an explicitly specified integer column instead of converting the column to float (GH2631)
- DatetimeIndex.unique now returns a DatetimeIndex with the same name and
- timezone instead of an array (GH2563)
New features¶
- MySQL support for database (contribution from Dan Allan)
HDFStore¶
You may need to upgrade your existing data files. Please visit the compatibility section in the main docs.
You can designate (and index) certain columns that you want to be able to perform queries on a table, by passing a list to data_columns
In [1870]: store = HDFStore('store.h5')
In [1871]: df = DataFrame(randn(8, 3), index=date_range('1/1/2000', periods=8),
......: columns=['A', 'B', 'C'])
......:
In [1872]: df['string'] = 'foo'
In [1873]: df.ix[4:6,'string'] = np.nan
In [1874]: df.ix[7:9,'string'] = 'bar'
In [1875]: df['string2'] = 'cool'
In [1876]: df
Out[1876]:
A B C string string2
2000-01-01 0.986719 1.550225 0.591428 foo cool
2000-01-02 0.919596 0.435997 -0.110372 foo cool
2000-01-03 1.097966 -0.789253 1.051532 foo cool
2000-01-04 1.647664 -0.837820 -1.708011 foo cool
2000-01-05 0.231848 0.358273 0.054422 NaN cool
2000-01-06 -0.104379 -0.910418 -0.607518 NaN cool
2000-01-07 -0.287767 -0.388098 -0.283159 foo cool
2000-01-08 -0.012229 1.043063 0.612015 bar cool
# on-disk operations
In [1877]: store.append('df', df, data_columns = ['B','C','string','string2'])
In [1878]: store.select('df',[ 'B > 0', 'string == foo' ])
Out[1878]:
A B C string string2
2000-01-01 0.986719 1.550225 0.591428 foo cool
2000-01-02 0.919596 0.435997 -0.110372 foo cool
# this is in-memory version of this type of selection
In [1879]: df[(df.B > 0) & (df.string == 'foo')]
Out[1879]:
A B C string string2
2000-01-01 0.986719 1.550225 0.591428 foo cool
2000-01-02 0.919596 0.435997 -0.110372 foo cool
Retrieving unique values in an indexable or data column.
In [1880]: store.unique('df','index')
Out[1880]:
array(['2000-01-01T02:00:00.000000000+0200',
'2000-01-02T02:00:00.000000000+0200',
'2000-01-03T02:00:00.000000000+0200',
'2000-01-04T02:00:00.000000000+0200',
'2000-01-05T02:00:00.000000000+0200',
'2000-01-06T02:00:00.000000000+0200',
'2000-01-07T02:00:00.000000000+0200',
'2000-01-08T02:00:00.000000000+0200'], dtype='datetime64[ns]')
In [1881]: store.unique('df','string')
Out[1881]: array(['foo', nan, 'bar'], dtype=object)
You can now store datetime64 in data columns
In [1882]: df_mixed = df.copy()
In [1883]: df_mixed['datetime64'] = Timestamp('20010102')
In [1884]: df_mixed.ix[3:4,['A','B']] = np.nan
In [1885]: store.append('df_mixed', df_mixed)
In [1886]: df_mixed1 = store.select('df_mixed')
In [1887]: df_mixed1
Out[1887]:
A B C string string2 datetime64
2000-01-01 0.986719 1.550225 0.591428 foo cool 2001-01-02 00:00:00
2000-01-02 0.919596 0.435997 -0.110372 foo cool 2001-01-02 00:00:00
2000-01-03 1.097966 -0.789253 1.051532 foo cool 2001-01-02 00:00:00
2000-01-04 NaN NaN -1.708011 foo cool 2001-01-02 00:00:00
2000-01-05 0.231848 0.358273 0.054422 NaN cool 2001-01-02 00:00:00
2000-01-06 -0.104379 -0.910418 -0.607518 NaN cool 2001-01-02 00:00:00
2000-01-07 -0.287767 -0.388098 -0.283159 foo cool 2001-01-02 00:00:00
2000-01-08 -0.012229 1.043063 0.612015 bar cool 2001-01-02 00:00:00
In [1888]: df_mixed1.get_dtype_counts()
Out[1888]:
datetime64[ns] 1
float64 3
object 2
dtype: int64
You can pass columns keyword to select to filter a list of the return columns, this is equivalent to passing a Term('columns',list_of_columns_to_filter)
In [1889]: store.select('df',columns = ['A','B'])
Out[1889]:
A B
2000-01-01 0.986719 1.550225
2000-01-02 0.919596 0.435997
2000-01-03 1.097966 -0.789253
2000-01-04 1.647664 -0.837820
2000-01-05 0.231848 0.358273
2000-01-06 -0.104379 -0.910418
2000-01-07 -0.287767 -0.388098
2000-01-08 -0.012229 1.043063
HDFStore now serializes multi-index dataframes when appending tables.
In [1890]: index = MultiIndex(levels=[['foo', 'bar', 'baz', 'qux'],
......: ['one', 'two', 'three']],
......: labels=[[0, 0, 0, 1, 1, 2, 2, 3, 3, 3],
......: [0, 1, 2, 0, 1, 1, 2, 0, 1, 2]],
......: names=['foo', 'bar'])
......:
In [1891]: df = DataFrame(np.random.randn(10, 3), index=index,
......: columns=['A', 'B', 'C'])
......:
In [1892]: df
Out[1892]:
A B C
foo bar
foo one 1.627605 0.670772 -0.611555
two 0.053425 -2.218806 0.634528
three 0.091848 -0.318810 0.950676
bar one -1.016290 -0.267508 0.115960
two -0.615949 -0.373060 0.276398
baz two -1.947432 -1.183044 -3.030491
three -1.055515 -0.177967 1.269136
qux one 0.668999 -0.234083 -0.254881
two -0.142302 1.291962 0.876700
three 1.704647 0.046376 0.158167
In [1893]: store.append('mi',df)
In [1894]: store.select('mi')
Out[1894]:
A B C
foo bar
foo one 1.627605 0.670772 -0.611555
two 0.053425 -2.218806 0.634528
three 0.091848 -0.318810 0.950676
bar one -1.016290 -0.267508 0.115960
two -0.615949 -0.373060 0.276398
baz two -1.947432 -1.183044 -3.030491
three -1.055515 -0.177967 1.269136
qux one 0.668999 -0.234083 -0.254881
two -0.142302 1.291962 0.876700
three 1.704647 0.046376 0.158167
# the levels are automatically included as data columns
In [1895]: store.select('mi', Term('foo=bar'))
Out[1895]:
A B C
foo bar
bar one -1.016290 -0.267508 0.115960
two -0.615949 -0.373060 0.276398
Multi-table creation via append_to_multiple and selection via select_as_multiple can create/select from multiple tables and return a combined result, by using where on a selector table.
In [1896]: df_mt = DataFrame(randn(8, 6), index=date_range('1/1/2000', periods=8),
......: columns=['A', 'B', 'C', 'D', 'E', 'F'])
......:
In [1897]: df_mt['foo'] = 'bar'
# you can also create the tables individually
In [1898]: store.append_to_multiple({ 'df1_mt' : ['A','B'], 'df2_mt' : None }, df_mt, selector = 'df1_mt')
In [1899]: store
Out[1899]:
<class 'pandas.io.pytables.HDFStore'>
File path: store.h5
/df frame_table (typ->appendable,nrows->8,ncols->5,indexers->[index],dc->[B,C,string,string2])
/df1_mt frame_table (typ->appendable,nrows->8,ncols->2,indexers->[index],dc->[A,B])
/df2_mt frame_table (typ->appendable,nrows->8,ncols->5,indexers->[index])
/df_mixed frame_table (typ->appendable,nrows->8,ncols->6,indexers->[index])
/mi frame_table (typ->appendable_multi,nrows->10,ncols->5,indexers->[index],dc->[bar,foo])
# indiviual tables were created
In [1900]: store.select('df1_mt')
Out[1900]:
A B
2000-01-01 1.503229 -0.335678
2000-01-02 -0.507624 -1.174443
2000-01-03 -0.323699 -1.378458
2000-01-04 0.345906 -1.778234
2000-01-05 1.247851 0.246737
2000-01-06 0.252915 -0.154549
2000-01-07 -0.778424 2.147255
2000-01-08 -0.058702 -1.297767
In [1901]: store.select('df2_mt')
Out[1901]:
C D E F foo
2000-01-01 0.157359 0.828373 0.860863 0.618679 bar
2000-01-02 0.191589 -0.243287 1.684079 -0.637764 bar
2000-01-03 -0.868599 1.916736 1.562215 0.133322 bar
2000-01-04 -1.223208 -0.480258 -0.285245 0.775414 bar
2000-01-05 1.454094 -1.166264 -0.560671 1.027488 bar
2000-01-06 0.181686 -0.268458 -0.124345 0.443256 bar
2000-01-07 -0.731309 0.281577 -0.417236 1.721160 bar
2000-01-08 0.871349 -0.177241 0.207366 2.592691 bar
# as a multiple
In [1902]: store.select_as_multiple(['df1_mt','df2_mt'], where = [ 'A>0','B>0' ], selector = 'df1_mt')
Out[1902]:
A B C D E F foo
2000-01-05 1.247851 0.246737 1.454094 -1.166264 -0.560671 1.027488 bar
Enhancements
- HDFStore now can read native PyTables table format tables
- You can pass nan_rep = 'my_nan_rep' to append, to change the default nan representation on disk (which converts to/from np.nan), this defaults to nan.
- You can pass index to append. This defaults to True. This will automagically create indicies on the indexables and data columns of the table
- You can pass chunksize=an integer to append, to change the writing chunksize (default is 50000). This will signficantly lower your memory usage on writing.
- You can pass expectedrows=an integer to the first append, to set the TOTAL number of expectedrows that PyTables will expected. This will optimize read/write performance.
- Select now supports passing start and stop to provide selection space limiting in selection.
- Greatly improved ISO8601 (e.g., yyyy-mm-dd) date parsing for file parsers (GH2698)
- Allow DataFrame.merge to handle combinatorial sizes too large for 64-bit integer (GH2690)
- Series now has unary negation (-series) and inversion (~series) operators (GH2686)
- DataFrame.plot now includes a logx parameter to change the x-axis to log scale (GH2327)
- Series arithmetic operators can now handle constant and ndarray input (GH2574)
- ExcelFile now takes a kind argument to specify the file type (GH2613)
- A faster implementation for Series.str methods (GH2602)
Bug Fixes
- HDFStore tables can now store float32 types correctly (cannot be mixed with float64 however)
- Fixed Google Analytics prefix when specifying request segment (GH2713).
- Function to reset Google Analytics token store so users can recover from improperly setup client secrets (GH2687).
- Fixed groupby bug resulting in segfault when passing in MultiIndex (GH2706)
- Fixed bug where passing a Series with datetime64 values into to_datetime results in bogus output values (GH2699)
- Fixed bug in pattern in HDFStore expressions when pattern is not a valid regex (GH2694)
- Fixed performance issues while aggregating boolean data (GH2692)
- When given a boolean mask key and a Series of new values, Series __setitem__ will now align the incoming values with the original Series (GH2686)
- Fixed MemoryError caused by performing counting sort on sorting MultiIndex levels with a very large number of combinatorial values (GH2684)
- Fixed bug that causes plotting to fail when the index is a DatetimeIndex with a fixed-offset timezone (GH2683)
- Corrected businessday subtraction logic when the offset is more than 5 bdays and the starting date is on a weekend (GH2680)
- Fixed C file parser behavior when the file has more columns than data (GH2668)
- Fixed file reader bug that misaligned columns with data in the presence of an implicit column and a specified usecols value
- DataFrames with numerical or datetime indices are now sorted prior to plotting (GH2609)
- Fixed DataFrame.from_records error when passed columns, index, but empty records (GH2633)
- Several bug fixed for Series operations when dtype is datetime64 (GH2689, GH2629, GH2626)
See the full release notes or issue tracker on GitHub for a complete list.
v0.10.0 (December 17, 2012)¶
This is a major release from 0.9.1 and includes many new features and enhancements along with a large number of bug fixes. There are also a number of important API changes that long-time pandas users should pay close attention to.
File parsing new features¶
The delimited file parsing engine (the guts of read_csv and read_table) has been rewritten from the ground up and now uses a fraction the amount of memory while parsing, while being 40% or more faster in most use cases (in some cases much faster).
There are also many new features:
- Much-improved Unicode handling via the encoding option.
- Column filtering (usecols)
- Dtype specification (dtype argument)
- Ability to specify strings to be recognized as True/False
- Ability to yield NumPy record arrays (as_recarray)
- High performance delim_whitespace option
- Decimal format (e.g. European format) specification
- Easier CSV dialect options: escapechar, lineterminator, quotechar, etc.
- More robust handling of many exceptional kinds of files observed in the wild
API changes¶
Deprecated DataFrame BINOP TimeSeries special case behavior
The default behavior of binary operations between a DataFrame and a Series has always been to align on the DataFrame’s columns and broadcast down the rows, except in the special case that the DataFrame contains time series. Since there are now method for each binary operator enabling you to specify how you want to broadcast, we are phasing out this special case (Zen of Python: Special cases aren’t special enough to break the rules). Here’s what I’m talking about:
In [1903]: import pandas as pd
In [1904]: df = pd.DataFrame(np.random.randn(6, 4),
......: index=pd.date_range('1/1/2000', periods=6))
......:
In [1905]: df
Out[1905]:
0 1 2 3
2000-01-01 0.423204 -0.006209 0.314186 0.363193
2000-01-02 0.196151 -1.598514 -0.843566 -0.353828
2000-01-03 0.516740 -2.335539 -0.715006 -0.399224
2000-01-04 0.798589 2.101702 -0.190649 0.595370
2000-01-05 -1.672567 0.786765 0.133175 -1.077265
2000-01-06 0.861068 1.982854 -1.059177 2.050701
# deprecated now
In [1906]: df - df[0]
Out[1906]:
0 1 2 3
2000-01-01 0 -0.429412 -0.109018 -0.060011
2000-01-02 0 -1.794664 -1.039717 -0.549979
2000-01-03 0 -2.852279 -1.231746 -0.915964
2000-01-04 0 1.303113 -0.989238 -0.203218
2000-01-05 0 2.459332 1.805743 0.595303
2000-01-06 0 1.121786 -1.920245 1.189633
# Change your code to
In [1907]: df.sub(df[0], axis=0) # align on axis 0 (rows)
Out[1907]:
0 1 2 3
2000-01-01 0 -0.429412 -0.109018 -0.060011
2000-01-02 0 -1.794664 -1.039717 -0.549979
2000-01-03 0 -2.852279 -1.231746 -0.915964
2000-01-04 0 1.303113 -0.989238 -0.203218
2000-01-05 0 2.459332 1.805743 0.595303
2000-01-06 0 1.121786 -1.920245 1.189633
You will get a deprecation warning in the 0.10.x series, and the deprecated functionality will be removed in 0.11 or later.
Altered resample default behavior
The default time series resample binning behavior of daily D and higher frequencies has been changed to closed='left', label='left'. Lower nfrequencies are unaffected. The prior defaults were causing a great deal of confusion for users, especially resampling data to daily frequency (which labeled the aggregated group with the end of the interval: the next day).
Note:
In [1908]: dates = pd.date_range('1/1/2000', '1/5/2000', freq='4h')
In [1909]: series = Series(np.arange(len(dates)), index=dates)
In [1910]: series
Out[1910]:
2000-01-01 00:00:00 0
2000-01-01 04:00:00 1
2000-01-01 08:00:00 2
2000-01-01 12:00:00 3
2000-01-01 16:00:00 4
2000-01-01 20:00:00 5
2000-01-02 00:00:00 6
2000-01-02 04:00:00 7
2000-01-02 08:00:00 8
2000-01-02 12:00:00 9
2000-01-02 16:00:00 10
2000-01-02 20:00:00 11
2000-01-03 00:00:00 12
2000-01-03 04:00:00 13
2000-01-03 08:00:00 14
2000-01-03 12:00:00 15
2000-01-03 16:00:00 16
2000-01-03 20:00:00 17
2000-01-04 00:00:00 18
2000-01-04 04:00:00 19
2000-01-04 08:00:00 20
2000-01-04 12:00:00 21
2000-01-04 16:00:00 22
2000-01-04 20:00:00 23
2000-01-05 00:00:00 24
Freq: 4H, dtype: int64
In [1911]: series.resample('D', how='sum')
Out[1911]:
2000-01-01 15
2000-01-02 51
2000-01-03 87
2000-01-04 123
2000-01-05 24
Freq: D, dtype: int64
# old behavior
In [1912]: series.resample('D', how='sum', closed='right', label='right')
Out[1912]:
2000-01-01 0
2000-01-02 21
2000-01-03 57
2000-01-04 93
2000-01-05 129
Freq: D, dtype: int64
- Infinity and negative infinity are no longer treated as NA by isnull and notnull. That they every were was a relic of early pandas. This behavior can be re-enabled globally by the mode.use_inf_as_null option:
In [1913]: s = pd.Series([1.5, np.inf, 3.4, -np.inf])
In [1914]: pd.isnull(s)
Out[1914]:
0 False
1 False
2 False
3 False
dtype: bool
In [1915]: s.fillna(0)
Out[1915]:
0 1.500000
1 inf
2 3.400000
3 -inf
dtype: float64
In [1916]: pd.set_option('use_inf_as_null', True)
In [1917]: pd.isnull(s)
Out[1917]:
0 False
1 True
2 False
3 True
dtype: bool
In [1918]: s.fillna(0)
Out[1918]:
0 1.5
1 0.0
2 3.4
3 0.0
dtype: float64
In [1919]: pd.reset_option('use_inf_as_null')
- Methods with the inplace option now all return None instead of the calling object. E.g. code written like df = df.fillna(0, inplace=True) may stop working. To fix, simply delete the unnecessary variable assignment.
- pandas.merge no longer sorts the group keys (sort=False) by default. This was done for performance reasons: the group-key sorting is often one of the more expensive parts of the computation and is often unnecessary.
- The default column names for a file with no header have been changed to the integers 0 through N - 1. This is to create consistency with the DataFrame constructor with no columns specified. The v0.9.0 behavior (names X0, X1, ...) can be reproduced by specifying prefix='X':
In [1920]: data= 'a,b,c\n1,Yes,2\n3,No,4'
In [1921]: print data
a,b,c
1,Yes,2
3,No,4
In [1922]: pd.read_csv(StringIO(data), header=None)
Out[1922]:
0 1 2
0 a b c
1 1 Yes 2
2 3 No 4
In [1923]: pd.read_csv(StringIO(data), header=None, prefix='X')
Out[1923]:
X0 X1 X2
0 a b c
1 1 Yes 2
2 3 No 4
- Values like 'Yes' and 'No' are not interpreted as boolean by default, though this can be controlled by new true_values and false_values arguments:
In [1924]: print data
a,b,c
1,Yes,2
3,No,4
In [1925]: pd.read_csv(StringIO(data))
Out[1925]:
a b c
0 1 Yes 2
1 3 No 4
In [1926]: pd.read_csv(StringIO(data), true_values=['Yes'], false_values=['No'])
Out[1926]:
a b c
0 1 True 2
1 3 False 4
- The file parsers will not recognize non-string values arising from a converter function as NA if passed in the na_values argument. It’s better to do post-processing using the replace function instead.
- Calling fillna on Series or DataFrame with no arguments is no longer valid code. You must either specify a fill value or an interpolation method:
In [1927]: s = Series([np.nan, 1., 2., np.nan, 4])
In [1928]: s
Out[1928]:
0 NaN
1 1
2 2
3 NaN
4 4
dtype: float64
In [1929]: s.fillna(0)
Out[1929]:
0 0
1 1
2 2
3 0
4 4
dtype: float64
In [1930]: s.fillna(method='pad')
Out[1930]:
0 NaN
1 1
2 2
3 2
4 4
dtype: float64
Convenience methods ffill and bfill have been added:
In [1931]: s.ffill()
Out[1931]:
0 NaN
1 1
2 2
3 2
4 4
dtype: float64
Series.apply will now operate on a returned value from the applied function, that is itself a series, and possibly upcast the result to a DataFrame
In [1932]: def f(x): ......: return Series([ x, x**2 ], index = ['x', 'x^2']) ......: In [1933]: s = Series(np.random.rand(5)) In [1934]: s Out[1934]: 0 0.209573 1 0.202737 2 0.014708 3 0.941394 4 0.332172 dtype: float64 In [1935]: s.apply(f) Out[1935]: x x^2 0 0.209573 0.043921 1 0.202737 0.041102 2 0.014708 0.000216 3 0.941394 0.886223 4 0.332172 0.110338
New API functions for working with pandas options (GH2097):
- get_option / set_option - get/set the value of an option. Partial names are accepted. - reset_option - reset one or more options to their default value. Partial names are accepted. - describe_option - print a description of one or more options. When called with no arguments. print all registered options.
Note: set_printoptions/ reset_printoptions are now deprecated (but functioning), the print options now live under “display.XYZ”. For example:
In [1936]: get_option("display.max_rows") Out[1936]: 60
to_string() methods now always return unicode strings (GH2224).
New features¶
Wide DataFrame Printing¶
Instead of printing the summary information, pandas now splits the string representation across multiple rows by default:
In [1937]: wide_frame = DataFrame(randn(5, 16))
In [1938]: wide_frame
Out[1938]:
0 1 2 3 4 5 6 \
0 1.554712 -0.931933 1.194806 -0.211196 -0.816904 -1.074726 -0.470691
1 -0.560488 -0.427787 -0.594425 -0.940300 -0.497396 -0.861299 0.217222
2 -0.224570 -0.325564 -0.830153 0.361426 1.080008 1.023402 1.417391
3 -0.453845 0.922367 1.107829 -0.463310 -1.138400 -1.284055 -0.600173
4 0.654298 -1.146232 1.144351 0.166619 0.147859 -1.333677 -0.171077
7 8 9 10 11 12 13 \
0 0.498441 0.833918 0.431463 0.447477 0.110952 -1.080534 0.831276
1 -0.785267 -0.960750 -0.137907 -0.844178 -1.435096 -0.092770 -1.739827
2 1.765283 0.684864 0.988679 0.301676 1.211569 2.847658 0.643408
3 0.341879 -0.420622 0.016883 -1.131983 -0.283679 -1.537059 0.163006
4 0.050424 -0.650290 -1.083796 -0.553609 -0.107442 -1.892957 0.460709
14 15
0 -1.678779 0.127673
1 1.366850 1.450803
2 1.887716 0.364659
3 -0.648131 -1.703280
4 0.253920 1.250457
The old behavior of printing out summary information can be achieved via the ‘expand_frame_repr’ print option:
In [1939]: pd.set_option('expand_frame_repr', False)
In [1940]: wide_frame
Out[1940]:
<class 'pandas.core.frame.DataFrame'>
Int64Index: 5 entries, 0 to 4
Data columns (total 16 columns):
0 5 non-null values
1 5 non-null values
2 5 non-null values
3 5 non-null values
4 5 non-null values
5 5 non-null values
6 5 non-null values
7 5 non-null values
8 5 non-null values
9 5 non-null values
10 5 non-null values
11 5 non-null values
12 5 non-null values
13 5 non-null values
14 5 non-null values
15 5 non-null values
dtypes: float64(16)
The width of each line can be changed via ‘line_width’ (80 by default):
In [1941]: pd.set_option('line_width', 40)
In [1942]: wide_frame
Out[1942]:
0 1 2 \
0 1.554712 -0.931933 1.194806
1 -0.560488 -0.427787 -0.594425
2 -0.224570 -0.325564 -0.830153
3 -0.453845 0.922367 1.107829
4 0.654298 -1.146232 1.144351
3 4 5 \
0 -0.211196 -0.816904 -1.074726
1 -0.940300 -0.497396 -0.861299
2 0.361426 1.080008 1.023402
3 -0.463310 -1.138400 -1.284055
4 0.166619 0.147859 -1.333677
6 7 8 \
0 -0.470691 0.498441 0.833918
1 0.217222 -0.785267 -0.960750
2 1.417391 1.765283 0.684864
3 -0.600173 0.341879 -0.420622
4 -0.171077 0.050424 -0.650290
9 10 11 \
0 0.431463 0.447477 0.110952
1 -0.137907 -0.844178 -1.435096
2 0.988679 0.301676 1.211569
3 0.016883 -1.131983 -0.283679
4 -1.083796 -0.553609 -0.107442
12 13 14 \
0 -1.080534 0.831276 -1.678779
1 -0.092770 -1.739827 1.366850
2 2.847658 0.643408 1.887716
3 -1.537059 0.163006 -0.648131
4 -1.892957 0.460709 0.253920
15
0 0.127673
1 1.450803
2 0.364659
3 -1.703280
4 1.250457
Updated PyTables Support¶
Docs for PyTables Table format & several enhancements to the api. Here is a taste of what to expect.
In [1943]: store = HDFStore('store.h5')
In [1944]: df = DataFrame(randn(8, 3), index=date_range('1/1/2000', periods=8),
......: columns=['A', 'B', 'C'])
......:
In [1945]: df
Out[1945]:
A B C
2000-01-01 0.526545 -0.877812 -0.624075
2000-01-02 -0.921519 2.133979 0.167893
2000-01-03 -0.480457 -0.626280 0.302336
2000-01-04 0.458588 0.788253 0.264381
2000-01-05 0.617429 -1.082697 -1.076447
2000-01-06 0.557384 -0.950833 0.479203
2000-01-07 -0.452393 -0.173608 0.050235
2000-01-08 -0.356023 0.190613 0.726404
# appending data frames
In [1946]: df1 = df[0:4]
In [1947]: df2 = df[4:]
In [1948]: store.append('df', df1)
In [1949]: store.append('df', df2)
In [1950]: store
Out[1950]:
<class 'pandas.io.pytables.HDFStore'>
File path: store.h5
/df frame_table (typ->appendable,nrows->8,ncols->3,indexers->[index])
# selecting the entire store
In [1951]: store.select('df')
Out[1951]:
A B C
2000-01-01 0.526545 -0.877812 -0.624075
2000-01-02 -0.921519 2.133979 0.167893
2000-01-03 -0.480457 -0.626280 0.302336
2000-01-04 0.458588 0.788253 0.264381
2000-01-05 0.617429 -1.082697 -1.076447
2000-01-06 0.557384 -0.950833 0.479203
2000-01-07 -0.452393 -0.173608 0.050235
2000-01-08 -0.356023 0.190613 0.726404
In [1952]: wp = Panel(randn(2, 5, 4), items=['Item1', 'Item2'],
......: major_axis=date_range('1/1/2000', periods=5),
......: minor_axis=['A', 'B', 'C', 'D'])
......:
In [1953]: wp
Out[1953]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 5 (major_axis) x 4 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 2000-01-01 00:00:00 to 2000-01-05 00:00:00
Minor_axis axis: A to D
# storing a panel
In [1954]: store.append('wp',wp)
# selecting via A QUERY
In [1955]: store.select('wp',
......: [ Term('major_axis>20000102'), Term('minor_axis', '=', ['A','B']) ])
......:
Out[1955]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 3 (major_axis) x 2 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 2000-01-03 00:00:00 to 2000-01-05 00:00:00
Minor_axis axis: A to B
# removing data from tables
In [1956]: store.remove('wp', [ 'major_axis', '>', wp.major_axis[3] ])
Out[1956]: 4
In [1957]: store.select('wp')
Out[1957]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 4 (major_axis) x 4 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 2000-01-01 00:00:00 to 2000-01-04 00:00:00
Minor_axis axis: A to D
# deleting a store
In [1958]: del store['df']
In [1959]: store
Out[1959]:
<class 'pandas.io.pytables.HDFStore'>
File path: store.h5
/wp wide_table (typ->appendable,nrows->16,ncols->2,indexers->[major_axis,minor_axis])
Enhancements
added ability to hierarchical keys
In [1960]: store.put('foo/bar/bah', df) In [1961]: store.append('food/orange', df) In [1962]: store.append('food/apple', df) In [1963]: store Out[1963]: <class 'pandas.io.pytables.HDFStore'> File path: store.h5 /wp wide_table (typ->appendable,nrows->16,ncols->2,indexers->[major_axis,minor_axis]) /food/apple frame_table (typ->appendable,nrows->8,ncols->3,indexers->[index]) /food/orange frame_table (typ->appendable,nrows->8,ncols->3,indexers->[index]) /foo/bar/bah frame (shape->[8,3]) # remove all nodes under this level In [1964]: store.remove('food') In [1965]: store Out[1965]: <class 'pandas.io.pytables.HDFStore'> File path: store.h5 /wp wide_table (typ->appendable,nrows->16,ncols->2,indexers->[major_axis,minor_axis]) /foo/bar/bah frame (shape->[8,3])
added mixed-dtype support!
In [1966]: df['string'] = 'string' In [1967]: df['int'] = 1 In [1968]: store.append('df',df) In [1969]: df1 = store.select('df') In [1970]: df1 Out[1970]: A B C string int 2000-01-01 0.526545 -0.877812 -0.624075 string 1 2000-01-02 -0.921519 2.133979 0.167893 string 1 2000-01-03 -0.480457 -0.626280 0.302336 string 1 2000-01-04 0.458588 0.788253 0.264381 string 1 2000-01-05 0.617429 -1.082697 -1.076447 string 1 2000-01-06 0.557384 -0.950833 0.479203 string 1 2000-01-07 -0.452393 -0.173608 0.050235 string 1 2000-01-08 -0.356023 0.190613 0.726404 string 1 In [1971]: df1.get_dtype_counts() Out[1971]: float64 3 int64 1 object 1 dtype: int64
performance improvments on table writing
support for arbitrarily indexed dimensions
SparseSeries now has a density property (GH2384)
enable Series.str.strip/lstrip/rstrip methods to take an input argument to strip arbitrary characters (GH2411)
implement value_vars in melt to limit values to certain columns and add melt to pandas namespace (GH2412)
Bug Fixes
- added Term method of specifying where conditions (GH1996).
- del store['df'] now call store.remove('df') for store deletion
- deleting of consecutive rows is much faster than before
- min_itemsize parameter can be specified in table creation to force a minimum size for indexing columns (the previous implementation would set the column size based on the first append)
- indexing support via create_table_index (requires PyTables >= 2.3) (GH698).
- appending on a store would fail if the table was not first created via put
- fixed issue with missing attributes after loading a pickled dataframe (GH2431)
- minor change to select and remove: require a table ONLY if where is also provided (and not None)
Compatibility
0.10 of HDFStore is backwards compatible for reading tables created in a prior version of pandas, however, query terms using the prior (undocumented) methodology are unsupported. You must read in the entire file and write it out using the new format to take advantage of the updates.
N Dimensional Panels (Experimental)¶
Adding experimental support for Panel4D and factory functions to create n-dimensional named panels. Docs for NDim. Here is a taste of what to expect.
In [1972]: p4d = Panel4D(randn(2, 2, 5, 4), ......: labels=['Label1','Label2'], ......: items=['Item1', 'Item2'], ......: major_axis=date_range('1/1/2000', periods=5), ......: minor_axis=['A', 'B', 'C', 'D']) ......: In [1973]: p4d Out[1973]: <class 'pandas.core.panelnd.Panel4D'> Dimensions: 2 (labels) x 2 (items) x 5 (major_axis) x 4 (minor_axis) Labels axis: Label1 to Label2 Items axis: Item1 to Item2 Major_axis axis: 2000-01-01 00:00:00 to 2000-01-05 00:00:00 Minor_axis axis: A to D
See the full release notes or issue tracker on GitHub for a complete list.
v0.9.1 (November 14, 2012)¶
This is a bugfix release from 0.9.0 and includes several new features and enhancements along with a large number of bug fixes. The new features include by-column sort order for DataFrame and Series, improved NA handling for the rank method, masking functions for DataFrame, and intraday time-series filtering for DataFrame.
New features¶
Series.sort, DataFrame.sort, and DataFrame.sort_index can now be specified in a per-column manner to support multiple sort orders (GH928)
In [1974]: df = DataFrame(np.random.randint(0, 2, (6, 3)), columns=['A', 'B', 'C']) In [1975]: df.sort(['A', 'B'], ascending=[1, 0]) Out[1975]: A B C 1 0 0 1 3 0 0 0 4 0 0 0 5 0 0 1 2 1 1 0 0 1 0 1DataFrame.rank now supports additional argument values for the na_option parameter so missing values can be assigned either the largest or the smallest rank (GH1508, GH2159)
In [1976]: df = DataFrame(np.random.randn(6, 3), columns=['A', 'B', 'C']) In [1977]: df.ix[2:4] = np.nan In [1978]: df.rank() Out[1978]: A B C 0 3 1 3 1 1 3 1 2 NaN NaN NaN 3 NaN NaN NaN 4 NaN NaN NaN 5 2 2 2 In [1979]: df.rank(na_option='top') Out[1979]: A B C 0 6 4 6 1 4 6 4 2 2 2 2 3 2 2 2 4 2 2 2 5 5 5 5 In [1980]: df.rank(na_option='bottom') Out[1980]: A B C 0 3 1 3 1 1 3 1 2 5 5 5 3 5 5 5 4 5 5 5 5 2 2 2DataFrame has new where and mask methods to select values according to a given boolean mask (GH2109, GH2151)
DataFrame currently supports slicing via a boolean vector the same length as the DataFrame (inside the []). The returned DataFrame has the same number of columns as the original, but is sliced on its index.
In [1981]: df = DataFrame(np.random.randn(5, 3), columns = ['A','B','C']) In [1982]: df Out[1982]: A B C 0 -0.531298 -0.065412 -1.043031 1 -0.658707 -0.866080 0.379561 2 -0.137358 0.006619 0.538026 3 -0.038056 -1.262660 0.151977 4 0.423176 2.545918 -1.070289 In [1983]: df[df['A'] > 0] Out[1983]: A B C 4 0.423176 2.545918 -1.070289If a DataFrame is sliced with a DataFrame based boolean condition (with the same size as the original DataFrame), then a DataFrame the same size (index and columns) as the original is returned, with elements that do not meet the boolean condition as NaN. This is accomplished via the new method DataFrame.where. In addition, where takes an optional other argument for replacement.
In [1984]: df[df>0] Out[1984]: A B C 0 NaN NaN NaN 1 NaN NaN 0.379561 2 NaN 0.006619 0.538026 3 NaN NaN 0.151977 4 0.423176 2.545918 NaN In [1985]: df.where(df>0) Out[1985]: A B C 0 NaN NaN NaN 1 NaN NaN 0.379561 2 NaN 0.006619 0.538026 3 NaN NaN 0.151977 4 0.423176 2.545918 NaN In [1986]: df.where(df>0,-df) Out[1986]: A B C 0 0.531298 0.065412 1.043031 1 0.658707 0.866080 0.379561 2 0.137358 0.006619 0.538026 3 0.038056 1.262660 0.151977 4 0.423176 2.545918 1.070289Furthermore, where now aligns the input boolean condition (ndarray or DataFrame), such that partial selection with setting is possible. This is analagous to partial setting via .ix (but on the contents rather than the axis labels)
In [1987]: df2 = df.copy() In [1988]: df2[ df2[1:4] > 0 ] = 3 In [1989]: df2 Out[1989]: A B C 0 -0.531298 -0.065412 -1.043031 1 -0.658707 -0.866080 3.000000 2 -0.137358 3.000000 3.000000 3 -0.038056 -1.262660 3.000000 4 0.423176 2.545918 -1.070289DataFrame.mask is the inverse boolean operation of where.
In [1990]: df.mask(df<=0) Out[1990]: A B C 0 NaN NaN NaN 1 NaN NaN 0.379561 2 NaN 0.006619 0.538026 3 NaN NaN 0.151977 4 0.423176 2.545918 NaNEnable referencing of Excel columns by their column names (GH1936)
In [1991]: xl = ExcelFile('data/test.xls') In [1992]: xl.parse('Sheet1', index_col=0, parse_dates=True, ......: parse_cols='A:D') ......: Out[1992]: A B C 2000-01-03 0.980269 3.685731 -0.364217 2000-01-04 1.047916 -0.041232 -0.161812 2000-01-05 0.498581 0.731168 -0.537677 2000-01-06 1.120202 1.567621 0.003641 2000-01-07 -0.487094 0.571455 -1.611639 2000-01-10 0.836649 0.246462 0.588543 2000-01-11 -0.157161 1.340307 1.195778Added option to disable pandas-style tick locators and formatters using series.plot(x_compat=True) or pandas.plot_params[‘x_compat’] = True (GH2205)
Existing TimeSeries methods at_time and between_time were added to DataFrame (GH2149)
DataFrame.dot can now accept ndarrays (GH2042)
DataFrame.drop now supports non-unique indexes (GH2101)
Panel.shift now supports negative periods (GH2164)
DataFrame now support unary ~ operator (GH2110)
API changes¶
Upsampling data with a PeriodIndex will result in a higher frequency TimeSeries that spans the original time window
In [1993]: prng = period_range('2012Q1', periods=2, freq='Q') In [1994]: s = Series(np.random.randn(len(prng)), prng) In [1995]: s.resample('M') Out[1995]: 2012-01 -1.411854 2012-02 NaN 2012-03 NaN 2012-04 0.026752 2012-05 NaN 2012-06 NaN Freq: M, dtype: float64Period.end_time now returns the last nanosecond in the time interval (GH2124, GH2125, GH1764)
In [1996]: p = Period('2012') In [1997]: p.end_time Out[1997]: <Timestamp: 2012-12-31 23:59:59.999999999>File parsers no longer coerce to float or bool for columns that have custom converters specified (GH2184)
In [1998]: data = 'A,B,C\n00001,001,5\n00002,002,6' In [1999]: from cStringIO import StringIO In [2000]: read_csv(StringIO(data), converters={'A' : lambda x: x.strip()}) Out[2000]: A B C 0 00001 1 5 1 00002 2 6
See the full release notes or issue tracker on GitHub for a complete list.
v0.9.0 (October 7, 2012)¶
This is a major release from 0.8.1 and includes several new features and enhancements along with a large number of bug fixes. New features include vectorized unicode encoding/decoding for Series.str, to_latex method to DataFrame, more flexible parsing of boolean values, and enabling the download of options data from Yahoo! Finance.
New features¶
- Add encode and decode for unicode handling to vectorized string processing methods in Series.str (GH1706)
- Add DataFrame.to_latex method (GH1735)
- Add convenient expanding window equivalents of all rolling_* ops (GH1785)
- Add Options class to pandas.io.data for fetching options data from Yahoo! Finance (GH1748, GH1739)
- More flexible parsing of boolean values (Yes, No, TRUE, FALSE, etc) (GH1691, GH1295)
- Add level parameter to Series.reset_index
- TimeSeries.between_time can now select times across midnight (GH1871)
- Series constructor can now handle generator as input (GH1679)
- DataFrame.dropna can now take multiple axes (tuple/list) as input (GH924)
- Enable skip_footer parameter in ExcelFile.parse (GH1843)
API changes¶
- The default column names when header=None and no columns names passed to functions like read_csv has changed to be more Pythonic and amenable to attribute access:
In [2001]: from StringIO import StringIO
In [2002]: data = '0,0,1\n1,1,0\n0,1,0'
In [2003]: df = read_csv(StringIO(data), header=None)
In [2004]: df
Out[2004]:
0 1 2
0 0 0 1
1 1 1 0
2 0 1 0
- Creating a Series from another Series, passing an index, will cause reindexing to happen inside rather than treating the Series like an ndarray. Technically improper usages like Series(df[col1], index=df[col2]) that worked before “by accident” (this was never intended) will lead to all NA Series in some cases. To be perfectly clear:
In [2005]: s1 = Series([1, 2, 3])
In [2006]: s1
Out[2006]:
0 1
1 2
2 3
dtype: int64
In [2007]: s2 = Series(s1, index=['foo', 'bar', 'baz'])
In [2008]: s2
Out[2008]:
foo NaN
bar NaN
baz NaN
dtype: float64
- Deprecated day_of_year API removed from PeriodIndex, use dayofyear (GH1723)
- Don’t modify NumPy suppress printoption to True at import time
- The internal HDF5 data arrangement for DataFrames has been transposed. Legacy files will still be readable by HDFStore (GH1834, GH1824)
- Legacy cruft removed: pandas.stats.misc.quantileTS
- Use ISO8601 format for Period repr: monthly, daily, and on down (GH1776)
- Empty DataFrame columns are now created as object dtype. This will prevent a class of TypeErrors that was occurring in code where the dtype of a column would depend on the presence of data or not (e.g. a SQL query having results) (GH1783)
- Setting parts of DataFrame/Panel using ix now aligns input Series/DataFrame (GH1630)
- first and last methods in GroupBy no longer drop non-numeric columns (GH1809)
- Resolved inconsistencies in specifying custom NA values in text parser. na_values of type dict no longer override default NAs unless keep_default_na is set to false explicitly (GH1657)
- DataFrame.dot will not do data alignment, and also work with Series (GH1915)
See the full release notes or issue tracker on GitHub for a complete list.
v0.8.1 (July 22, 2012)¶
This release includes a few new features, performance enhancements, and over 30 bug fixes from 0.8.0. New features include notably NA friendly string processing functionality and a series of new plot types and options.
New features¶
- Add vectorized string processing methods accessible via Series.str (GH620)
- Add option to disable adjustment in EWMA (GH1584)
- Radviz plot (GH1566)
- Parallel coordinates plot
- Bootstrap plot
- Per column styles and secondary y-axis plotting (GH1559)
- New datetime converters millisecond plotting (GH1599)
- Add option to disable “sparse” display of hierarchical indexes (GH1538)
- Series/DataFrame’s set_index method can append levels to an existing Index/MultiIndex (GH1569, GH1577)
Performance improvements¶
- Improved implementation of rolling min and max (thanks to Bottleneck !)
- Add accelerated 'median' GroupBy option (GH1358)
- Significantly improve the performance of parsing ISO8601-format date strings with DatetimeIndex or to_datetime (GH1571)
- Improve the performance of GroupBy on single-key aggregations and use with Categorical types
- Significant datetime parsing performance improvments
v0.8.0 (June 29, 2012)¶
This is a major release from 0.7.3 and includes extensive work on the time series handling and processing infrastructure as well as a great deal of new functionality throughout the library. It includes over 700 commits from more than 20 distinct authors. Most pandas 0.7.3 and earlier users should not experience any issues upgrading, but due to the migration to the NumPy datetime64 dtype, there may be a number of bugs and incompatibilities lurking. Lingering incompatibilities will be fixed ASAP in a 0.8.1 release if necessary. See the full release notes or issue tracker on GitHub for a complete list.
Support for non-unique indexes¶
All objects can now work with non-unique indexes. Data alignment / join operations work according to SQL join semantics (including, if application, index duplication in many-to-many joins)
NumPy datetime64 dtype and 1.6 dependency¶
Time series data are now represented using NumPy’s datetime64 dtype; thus, pandas 0.8.0 now requires at least NumPy 1.6. It has been tested and verified to work with the development version (1.7+) of NumPy as well which includes some significant user-facing API changes. NumPy 1.6 also has a number of bugs having to do with nanosecond resolution data, so I recommend that you steer clear of NumPy 1.6’s datetime64 API functions (though limited as they are) and only interact with this data using the interface that pandas provides.
See the end of the 0.8.0 section for a “porting” guide listing potential issues for users migrating legacy codebases from pandas 0.7 or earlier to 0.8.0.
Bug fixes to the 0.7.x series for legacy NumPy < 1.6 users will be provided as they arise. There will be no more further development in 0.7.x beyond bug fixes.
Time series changes and improvements¶
Note
With this release, legacy scikits.timeseries users should be able to port their code to use pandas.
Note
See documentation for overview of pandas timeseries API.
- New datetime64 representation speeds up join operations and data alignment, reduces memory usage, and improve serialization / deserialization performance significantly over datetime.datetime
- High performance and flexible resample method for converting from high-to-low and low-to-high frequency. Supports interpolation, user-defined aggregation functions, and control over how the intervals and result labeling are defined. A suite of high performance Cython/C-based resampling functions (including Open-High-Low-Close) have also been implemented.
- Revamp of frequency aliases and support for frequency shortcuts like ‘15min’, or ‘1h30min’
- New DatetimeIndex class supports both fixed frequency and irregular time series. Replaces now deprecated DateRange class
- New PeriodIndex and Period classes for representing time spans and performing calendar logic, including the 12 fiscal quarterly frequencies <timeseries.quarterly>. This is a partial port of, and a substantial enhancement to, elements of the scikits.timeseries codebase. Support for conversion between PeriodIndex and DatetimeIndex
- New Timestamp data type subclasses datetime.datetime, providing the same interface while enabling working with nanosecond-resolution data. Also provides easy time zone conversions.
- Enhanced support for time zones. Add tz_convert and tz_lcoalize methods to TimeSeries and DataFrame. All timestamps are stored as UTC; Timestamps from DatetimeIndex objects with time zone set will be localized to localtime. Time zone conversions are therefore essentially free. User needs to know very little about pytz library now; only time zone names as as strings are required. Time zone-aware timestamps are equal if and only if their UTC timestamps match. Operations between time zone-aware time series with different time zones will result in a UTC-indexed time series.
- Time series string indexing conveniences / shortcuts: slice years, year and month, and index values with strings
- Enhanced time series plotting; adaptation of scikits.timeseries matplotlib-based plotting code
- New date_range, bdate_range, and period_range factory functions
- Robust frequency inference function infer_freq and inferred_freq property of DatetimeIndex, with option to infer frequency on construction of DatetimeIndex
- to_datetime function efficiently parses array of strings to DatetimeIndex. DatetimeIndex will parse array or list of strings to datetime64
- Optimized support for datetime64-dtype data in Series and DataFrame columns
- New NaT (Not-a-Time) type to represent NA in timestamp arrays
- Optimize Series.asof for looking up “as of” values for arrays of timestamps
- Milli, Micro, Nano date offset objects
- Can index time series with datetime.time objects to select all data at particular time of day (TimeSeries.at_time) or between two times (TimeSeries.between_time)
- Add tshift method for leading/lagging using the frequency (if any) of the index, as opposed to a naive lead/lag using shift
Other new features¶
- New cut and qcut functions (like R’s cut function) for computing a categorical variable from a continuous variable by binning values either into value-based (cut) or quantile-based (qcut) bins
- Rename Factor to Categorical and add a number of usability features
- Add limit argument to fillna/reindex
- More flexible multiple function application in GroupBy, and can pass list (name, function) tuples to get result in particular order with given names
- Add flexible replace method for efficiently substituting values
- Enhanced read_csv/read_table for reading time series data and converting multiple columns to dates
- Add comments option to parser functions: read_csv, etc.
- Add :ref`dayfirst <io.dayfirst>` option to parser functions for parsing international DD/MM/YYYY dates
- Allow the user to specify the CSV reader dialect to control quoting etc.
- Handling thousands separators in read_csv to improve integer parsing.
- Enable unstacking of multiple levels in one shot. Alleviate pivot_table bugs (empty columns being introduced)
- Move to klib-based hash tables for indexing; better performance and less memory usage than Python’s dict
- Add first, last, min, max, and prod optimized GroupBy functions
- New ordered_merge function
- Add flexible comparison instance methods eq, ne, lt, gt, etc. to DataFrame, Series
- Improve scatter_matrix plotting function and add histogram or kernel density estimates to diagonal
- Add ‘kde’ plot option for density plots
- Support for converting DataFrame to R data.frame through rpy2
- Improved support for complex numbers in Series and DataFrame
- Add pct_change method to all data structures
- Add max_colwidth configuration option for DataFrame console output
- Interpolate Series values using index values
- Can select multiple columns from GroupBy
- Add update methods to Series/DataFrame for updating values in place
- Add any and all method to DataFrame
New plotting methods¶
Series.plot now supports a secondary_y option:
In [2009]: plt.figure()
Out[2009]: <matplotlib.figure.Figure at 0x198bd550>
In [2010]: fx['FR'].plot(style='g')
Out[2010]: <matplotlib.axes.AxesSubplot at 0x198bdbd0>
In [2011]: fx['IT'].plot(style='k--', secondary_y=True)
Out[2011]: <matplotlib.axes.AxesSubplot at 0x198e4390>
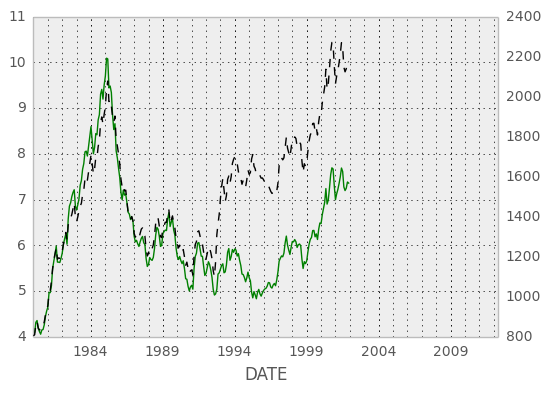
Vytautas Jancauskas, the 2012 GSOC participant, has added many new plot types. For example, 'kde' is a new option:
In [2012]: s = Series(np.concatenate((np.random.randn(1000),
......: np.random.randn(1000) * 0.5 + 3)))
......:
In [2013]: plt.figure()
Out[2013]: <matplotlib.figure.Figure at 0x19f60b50>
In [2014]: s.hist(normed=True, alpha=0.2)
Out[2014]: <matplotlib.axes.AxesSubplot at 0x18df4450>
In [2015]: s.plot(kind='kde')
Out[2015]: <matplotlib.axes.AxesSubplot at 0x18df4450>
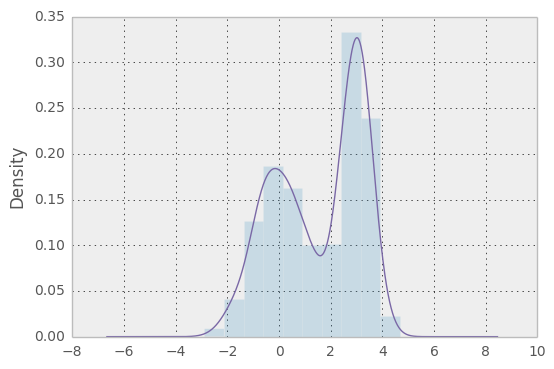
See the plotting page for much more.
Other API changes¶
- Deprecation of offset, time_rule, and timeRule arguments names in time series functions. Warnings will be printed until pandas 0.9 or 1.0.
Potential porting issues for pandas <= 0.7.3 users¶
The major change that may affect you in pandas 0.8.0 is that time series indexes use NumPy’s datetime64 data type instead of dtype=object arrays of Python’s built-in datetime.datetime objects. DateRange has been replaced by DatetimeIndex but otherwise behaved identically. But, if you have code that converts DateRange or Index objects that used to contain datetime.datetime values to plain NumPy arrays, you may have bugs lurking with code using scalar values because you are handing control over to NumPy:
In [2016]: import datetime
In [2017]: rng = date_range('1/1/2000', periods=10)
In [2018]: rng[5]
Out[2018]: <Timestamp: 2000-01-06 00:00:00>
In [2019]: isinstance(rng[5], datetime.datetime)
Out[2019]: True
In [2020]: rng_asarray = np.asarray(rng)
In [2021]: scalar_val = rng_asarray[5]
In [2022]: type(scalar_val)
Out[2022]: numpy.datetime64
pandas’s Timestamp object is a subclass of datetime.datetime that has nanosecond support (the nanosecond field store the nanosecond value between 0 and 999). It should substitute directly into any code that used datetime.datetime values before. Thus, I recommend not casting DatetimeIndex to regular NumPy arrays.
If you have code that requires an array of datetime.datetime objects, you have a couple of options. First, the asobject property of DatetimeIndex produces an array of Timestamp objects:
In [2023]: stamp_array = rng.asobject
In [2024]: stamp_array
Out[2024]: Index([2000-01-01 00:00:00, 2000-01-02 00:00:00, 2000-01-03 00:00:00, 2000-01-04 00:00:00, 2000-01-05 00:00:00, 2000-01-06 00:00:00, 2000-01-07 00:00:00, 2000-01-08 00:00:00, 2000-01-09 00:00:00, 2000-01-10 00:00:00], dtype=object)
In [2025]: stamp_array[5]
Out[2025]: <Timestamp: 2000-01-06 00:00:00>
To get an array of proper datetime.datetime objects, use the to_pydatetime method:
In [2026]: dt_array = rng.to_pydatetime()
In [2027]: dt_array
Out[2027]:
array([datetime.datetime(2000, 1, 1, 0, 0),
datetime.datetime(2000, 1, 2, 0, 0),
datetime.datetime(2000, 1, 3, 0, 0),
datetime.datetime(2000, 1, 4, 0, 0),
datetime.datetime(2000, 1, 5, 0, 0),
datetime.datetime(2000, 1, 6, 0, 0),
datetime.datetime(2000, 1, 7, 0, 0),
datetime.datetime(2000, 1, 8, 0, 0),
datetime.datetime(2000, 1, 9, 0, 0),
datetime.datetime(2000, 1, 10, 0, 0)], dtype=object)
In [2028]: dt_array[5]
Out[2028]: datetime.datetime(2000, 1, 6, 0, 0)
matplotlib knows how to handle datetime.datetime but not Timestamp objects. While I recommend that you plot time series using TimeSeries.plot, you can either use to_pydatetime or register a converter for the Timestamp type. See matplotlib documentation for more on this.
Warning
There are bugs in the user-facing API with the nanosecond datetime64 unit in NumPy 1.6. In particular, the string version of the array shows garbage values, and conversion to dtype=object is similarly broken.
In [2029]: rng = date_range('1/1/2000', periods=10)
In [2030]: rng
Out[2030]:
<class 'pandas.tseries.index.DatetimeIndex'>
[2000-01-01 00:00:00, ..., 2000-01-10 00:00:00]
Length: 10, Freq: D, Timezone: None
In [2031]: np.asarray(rng)
Out[2031]:
array(['2000-01-01T02:00:00.000000000+0200',
'2000-01-02T02:00:00.000000000+0200',
'2000-01-03T02:00:00.000000000+0200',
'2000-01-04T02:00:00.000000000+0200',
'2000-01-05T02:00:00.000000000+0200',
'2000-01-06T02:00:00.000000000+0200',
'2000-01-07T02:00:00.000000000+0200',
'2000-01-08T02:00:00.000000000+0200',
'2000-01-09T02:00:00.000000000+0200',
'2000-01-10T02:00:00.000000000+0200'], dtype='datetime64[ns]')
In [2032]: converted = np.asarray(rng, dtype=object)
In [2033]: converted[5]
Out[2033]: 947116800000000000L
Trust me: don’t panic. If you are using NumPy 1.6 and restrict your interaction with datetime64 values to pandas’s API you will be just fine. There is nothing wrong with the data-type (a 64-bit integer internally); all of the important data processing happens in pandas and is heavily tested. I strongly recommend that you do not work directly with datetime64 arrays in NumPy 1.6 and only use the pandas API.
Support for non-unique indexes: In the latter case, you may have code inside a try:... catch: block that failed due to the index not being unique. In many cases it will no longer fail (some method like append still check for uniqueness unless disabled). However, all is not lost: you can inspect index.is_unique and raise an exception explicitly if it is False or go to a different code branch.
v.0.7.3 (April 12, 2012)¶
This is a minor release from 0.7.2 and fixes many minor bugs and adds a number of nice new features. There are also a couple of API changes to note; these should not affect very many users, and we are inclined to call them “bug fixes” even though they do constitute a change in behavior. See the full release notes or issue tracker on GitHub for a complete list.
New features¶
- New fixed width file reader, read_fwf
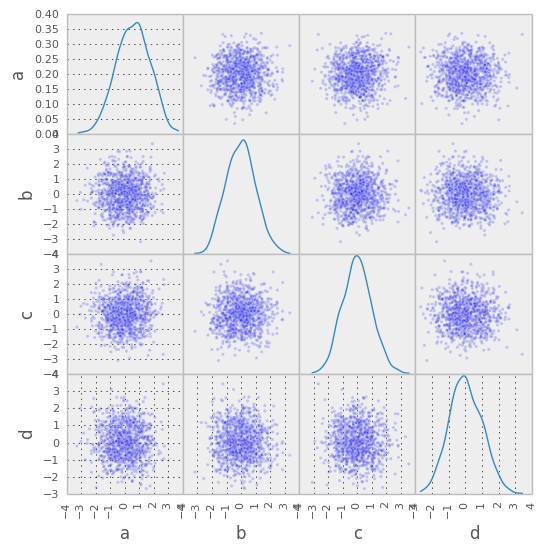
- New scatter_matrix function for making a scatter plot matrix
from pandas.tools.plotting import scatter_matrix
scatter_matrix(df, alpha=0.2)
- Add stacked argument to Series and DataFrame’s plot method for stacked bar plots.
df.plot(kind='bar', stacked=True)
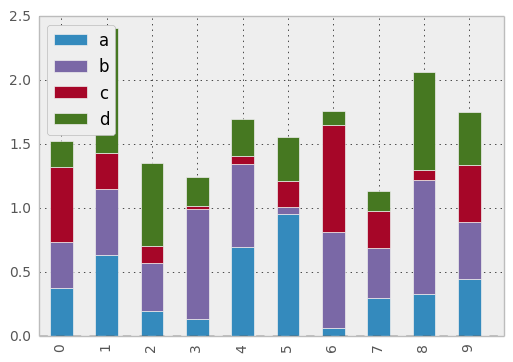
df.plot(kind='barh', stacked=True)
- Add log x and y scaling options to DataFrame.plot and Series.plot
- Add kurt methods to Series and DataFrame for computing kurtosis
NA Boolean Comparison API Change¶
Reverted some changes to how NA values (represented typically as NaN or None) are handled in non-numeric Series:
In [2034]: series = Series(['Steve', np.nan, 'Joe'])
In [2035]: series == 'Steve'
Out[2035]:
0 True
1 False
2 False
dtype: bool
In [2036]: series != 'Steve'
Out[2036]:
0 False
1 True
2 True
dtype: bool
In comparisons, NA / NaN will always come through as False except with != which is True. Be very careful with boolean arithmetic, especially negation, in the presence of NA data. You may wish to add an explicit NA filter into boolean array operations if you are worried about this:
In [2037]: mask = series == 'Steve'
In [2038]: series[mask & series.notnull()]
Out[2038]:
0 Steve
dtype: object
While propagating NA in comparisons may seem like the right behavior to some users (and you could argue on purely technical grounds that this is the right thing to do), the evaluation was made that propagating NA everywhere, including in numerical arrays, would cause a large amount of problems for users. Thus, a “practicality beats purity” approach was taken. This issue may be revisited at some point in the future.
Other API Changes¶
When calling apply on a grouped Series, the return value will also be a Series, to be more consistent with the groupby behavior with DataFrame:
In [2039]: df = DataFrame({'A' : ['foo', 'bar', 'foo', 'bar',
......: 'foo', 'bar', 'foo', 'foo'],
......: 'B' : ['one', 'one', 'two', 'three',
......: 'two', 'two', 'one', 'three'],
......: 'C' : np.random.randn(8), 'D' : np.random.randn(8)})
......:
In [2040]: df
Out[2040]:
A B C D
0 foo one 0.565554 0.028444
1 bar one -0.040251 0.418069
2 foo two -0.492753 -0.165726
3 bar three -0.834185 -0.610824
4 foo two -1.235635 0.130725
5 bar two 0.234011 -0.366952
6 foo one 1.402164 -0.242016
7 foo three -0.803155 0.318309
In [2041]: grouped = df.groupby('A')['C']
In [2042]: grouped.describe()
Out[2042]:
A
bar count 3.000000
mean -0.213475
std 0.554766
min -0.834185
25% -0.437218
50% -0.040251
75% 0.096880
max 0.234011
foo count 5.000000
mean -0.112765
std 1.076684
min -1.235635
25% -0.803155
50% -0.492753
75% 0.565554
max 1.402164
dtype: float64
In [2043]: grouped.apply(lambda x: x.order()[-2:]) # top 2 values
Out[2043]:
A
bar 1 -0.040251
5 0.234011
foo 0 0.565554
6 1.402164
dtype: float64
v.0.7.2 (March 16, 2012)¶
This release targets bugs in 0.7.1, and adds a few minor features.
New features¶
- Add additional tie-breaking methods in DataFrame.rank (GH874)
- Add ascending parameter to rank in Series, DataFrame (GH875)
- Add coerce_float option to DataFrame.from_records (GH893)
- Add sort_columns parameter to allow unsorted plots (GH918)
- Enable column access via attributes on GroupBy (GH882)
- Can pass dict of values to DataFrame.fillna (GH661)
- Can select multiple hierarchical groups by passing list of values in .ix (GH134)
- Add axis option to DataFrame.fillna (GH174)
- Add level keyword to drop for dropping values from a level (GH159)
v.0.7.1 (February 29, 2012)¶
This release includes a few new features and addresses over a dozen bugs in 0.7.0.
New features¶
- Add to_clipboard function to pandas namespace for writing objects to the system clipboard (GH774)
- Add itertuples method to DataFrame for iterating through the rows of a dataframe as tuples (GH818)
- Add ability to pass fill_value and method to DataFrame and Series align method (GH806, GH807)
- Add fill_value option to reindex, align methods (GH784)
- Enable concat to produce DataFrame from Series (GH787)
- Add between method to Series (GH802)
- Add HTML representation hook to DataFrame for the IPython HTML notebook (GH773)
- Support for reading Excel 2007 XML documents using openpyxl
v.0.7.0 (February 9, 2012)¶
New features¶
- New unified merge function for efficiently performing full gamut of database / relational-algebra operations. Refactored existing join methods to use the new infrastructure, resulting in substantial performance gains (GH220, GH249, GH267)
- New unified concatenation function for concatenating Series, DataFrame or Panel objects along an axis. Can form union or intersection of the other axes. Improves performance of Series.append and DataFrame.append (GH468, GH479, GH273)
- Can pass multiple DataFrames to DataFrame.append to concatenate (stack) and multiple Series to Series.append too
- Can pass list of dicts (e.g., a list of JSON objects) to DataFrame constructor (GH526)
- You can now set multiple columns in a DataFrame via __getitem__, useful for transformation (GH342)
- Handle differently-indexed output values in DataFrame.apply (GH498)
In [2044]: df = DataFrame(randn(10, 4))
In [2045]: df.apply(lambda x: x.describe())
Out[2045]:
0 1 2 3
count 10.000000 10.000000 10.000000 10.000000
mean -0.473881 -0.596460 0.127205 0.168917
std 1.266731 0.566807 0.888104 0.856847
min -3.152616 -1.398390 -1.428126 -1.353873
25% -1.005760 -1.151049 0.059401 -0.302776
50% -0.411972 -0.458980 0.180852 0.267014
75% 0.087190 -0.131078 0.378182 0.893358
max 1.482459 0.110916 1.352172 1.163741
- Add reorder_levels method to Series and DataFrame (PR534)
- Add dict-like get function to DataFrame and Panel (PR521)
- Add DataFrame.iterrows method for efficiently iterating through the rows of a DataFrame
- Add DataFrame.to_panel with code adapted from LongPanel.to_long
- Add reindex_axis method added to DataFrame
- Add level option to binary arithmetic functions on DataFrame and Series
- Add level option to the reindex and align methods on Series and DataFrame for broadcasting values across a level (GH542, PR552, others)
- Add attribute-based item access to Panel and add IPython completion (PR563)
- Add logy option to Series.plot for log-scaling on the Y axis
- Add index and header options to DataFrame.to_string
- Can pass multiple DataFrames to DataFrame.join to join on index (GH115)
- Can pass multiple Panels to Panel.join (GH115)
- Added justify argument to DataFrame.to_string to allow different alignment of column headers
- Add sort option to GroupBy to allow disabling sorting of the group keys for potential speedups (GH595)
- Can pass MaskedArray to Series constructor (PR563)
- Add Panel item access via attributes and IPython completion (GH554)
- Implement DataFrame.lookup, fancy-indexing analogue for retrieving values given a sequence of row and column labels (GH338)
- Can pass a list of functions to aggregate with groupby on a DataFrame, yielding an aggregated result with hierarchical columns (GH166)
- Can call cummin and cummax on Series and DataFrame to get cumulative minimum and maximum, respectively (GH647)
- value_range added as utility function to get min and max of a dataframe (GH288)
- Added encoding argument to read_csv, read_table, to_csv and from_csv for non-ascii text (GH717)
- Added abs method to pandas objects
- Added crosstab function for easily computing frequency tables
- Added isin method to index objects
- Added level argument to xs method of DataFrame.
API Changes to integer indexing¶
One of the potentially riskiest API changes in 0.7.0, but also one of the most important, was a complete review of how integer indexes are handled with regard to label-based indexing. Here is an example:
In [2046]: s = Series(randn(10), index=range(0, 20, 2))
In [2047]: s
Out[2047]:
0 0.162121
2 0.581910
4 0.305402
6 0.578765
8 -0.369912
10 -0.284429
12 -0.947215
14 -0.212794
16 -0.677290
18 -0.791236
dtype: float64
In [2048]: s[0]
Out[2048]: 0.16212102647561361
In [2049]: s[2]
Out[2049]: 0.58191028914602694
In [2050]: s[4]
Out[2050]: 0.30540242017176711
This is all exactly identical to the behavior before. However, if you ask for a key not contained in the Series, in versions 0.6.1 and prior, Series would fall back on a location-based lookup. This now raises a KeyError:
In [2]: s[1]
KeyError: 1
This change also has the same impact on DataFrame:
In [3]: df = DataFrame(randn(8, 4), index=range(0, 16, 2))
In [4]: df
0 1 2 3
0 0.88427 0.3363 -0.1787 0.03162
2 0.14451 -0.1415 0.2504 0.58374
4 -1.44779 -0.9186 -1.4996 0.27163
6 -0.26598 -2.4184 -0.2658 0.11503
8 -0.58776 0.3144 -0.8566 0.61941
10 0.10940 -0.7175 -1.0108 0.47990
12 -1.16919 -0.3087 -0.6049 -0.43544
14 -0.07337 0.3410 0.0424 -0.16037
In [5]: df.ix[3]
KeyError: 3
In order to support purely integer-based indexing, the following methods have been added:
| Method | Description |
|---|---|
| Series.iget_value(i) | Retrieve value stored at location i |
| Series.iget(i) | Alias for iget_value |
| DataFrame.irow(i) | Retrieve the i-th row |
| DataFrame.icol(j) | Retrieve the j-th column |
| DataFrame.iget_value(i, j) | Retrieve the value at row i and column j |
API tweaks regarding label-based slicing¶
Label-based slicing using ix now requires that the index be sorted (monotonic) unless both the start and endpoint are contained in the index:
In [2051]: s = Series(randn(6), index=list('gmkaec'))
In [2052]: s
Out[2052]:
g 0.550334
m -0.631881
k 0.388663
a -0.064094
e -0.059266
c 0.956671
dtype: float64
Then this is OK:
In [2053]: s.ix['k':'e']
Out[2053]:
k 0.388663
a -0.064094
e -0.059266
dtype: float64
But this is not:
In [12]: s.ix['b':'h']
KeyError 'b'
If the index had been sorted, the “range selection” would have been possible:
In [2054]: s2 = s.sort_index()
In [2055]: s2
Out[2055]:
a -0.064094
c 0.956671
e -0.059266
g 0.550334
k 0.388663
m -0.631881
dtype: float64
In [2056]: s2.ix['b':'h']
Out[2056]:
c 0.956671
e -0.059266
g 0.550334
dtype: float64
Changes to Series [] operator¶
As as notational convenience, you can pass a sequence of labels or a label slice to a Series when getting and setting values via [] (i.e. the __getitem__ and __setitem__ methods). The behavior will be the same as passing similar input to ix except in the case of integer indexing:
In [2057]: s = Series(randn(6), index=list('acegkm'))
In [2058]: s
Out[2058]:
a -0.131986
c -0.279014
e -1.444146
g -1.074302
k 0.032490
m -0.205971
dtype: float64
In [2059]: s[['m', 'a', 'c', 'e']]
Out[2059]:
m -0.205971
a -0.131986
c -0.279014
e -1.444146
dtype: float64
In [2060]: s['b':'l']
Out[2060]:
c -0.279014
e -1.444146
g -1.074302
k 0.032490
dtype: float64
In [2061]: s['c':'k']
Out[2061]:
c -0.279014
e -1.444146
g -1.074302
k 0.032490
dtype: float64
In the case of integer indexes, the behavior will be exactly as before (shadowing ndarray):
In [2062]: s = Series(randn(6), index=range(0, 12, 2))
In [2063]: s[[4, 0, 2]]
Out[2063]:
4 2.326354
0 -1.683462
2 -0.434042
dtype: float64
In [2064]: s[1:5]
Out[2064]:
2 -0.434042
4 2.326354
6 -1.941687
8 0.575285
dtype: float64
If you wish to do indexing with sequences and slicing on an integer index with label semantics, use ix.
Other API Changes¶
- The deprecated LongPanel class has been completely removed
- If Series.sort is called on a column of a DataFrame, an exception will now be raised. Before it was possible to accidentally mutate a DataFrame’s column by doing df[col].sort() instead of the side-effect free method df[col].order() (GH316)
- Miscellaneous renames and deprecations which will (harmlessly) raise FutureWarning
- drop added as an optional parameter to DataFrame.reset_index (GH699)
Performance improvements¶
- Cythonized GroupBy aggregations no longer presort the data, thus achieving a significant speedup (GH93). GroupBy aggregations with Python functions significantly sped up by clever manipulation of the ndarray data type in Cython (GH496).
- Better error message in DataFrame constructor when passed column labels don’t match data (GH497)
- Substantially improve performance of multi-GroupBy aggregation when a Python function is passed, reuse ndarray object in Cython (GH496)
- Can store objects indexed by tuples and floats in HDFStore (GH492)
- Don’t print length by default in Series.to_string, add length option (GH489)
- Improve Cython code for multi-groupby to aggregate without having to sort the data (GH93)
- Improve MultiIndex reindexing speed by storing tuples in the MultiIndex, test for backwards unpickling compatibility
- Improve column reindexing performance by using specialized Cython take function
- Further performance tweaking of Series.__getitem__ for standard use cases
- Avoid Index dict creation in some cases (i.e. when getting slices, etc.), regression from prior versions
- Friendlier error message in setup.py if NumPy not installed
- Use common set of NA-handling operations (sum, mean, etc.) in Panel class also (GH536)
- Default name assignment when calling reset_index on DataFrame with a regular (non-hierarchical) index (GH476)
- Use Cythonized groupers when possible in Series/DataFrame stat ops with level parameter passed (GH545)
- Ported skiplist data structure to C to speed up rolling_median by about 5-10x in most typical use cases (GH374)
v.0.6.1 (December 13, 2011)¶
New features¶
- Can append single rows (as Series) to a DataFrame
- Add Spearman and Kendall rank correlation options to Series.corr and DataFrame.corr (GH428)
- Added get_value and set_value methods to Series, DataFrame, and Panel for very low-overhead access (>2x faster in many cases) to scalar elements (GH437, GH438). set_value is capable of producing an enlarged object.
- Add PyQt table widget to sandbox (PR435)
- DataFrame.align can accept Series arguments and an axis option (GH461)
- Implement new SparseArray and SparseList data structures. SparseSeries now derives from SparseArray (GH463)
- Better console printing options (PR453)
- Implement fast data ranking for Series and DataFrame, fast versions of scipy.stats.rankdata (GH428)
- Implement DataFrame.from_items alternate constructor (GH444)
- DataFrame.convert_objects method for inferring better dtypes for object columns (GH302)
- Add rolling_corr_pairwise function for computing Panel of correlation matrices (GH189)
- Add margins option to pivot_table for computing subgroup aggregates (GH114)
- Add Series.from_csv function (PR482)
- Can pass DataFrame/DataFrame and DataFrame/Series to rolling_corr/rolling_cov (GH #462)
- MultiIndex.get_level_values can accept the level name
Performance improvements¶
- Improve memory usage of DataFrame.describe (do not copy data unnecessarily) (PR #425)
- Optimize scalar value lookups in the general case by 25% or more in Series and DataFrame
- Fix performance regression in cross-sectional count in DataFrame, affecting DataFrame.dropna speed
- Column deletion in DataFrame copies no data (computes views on blocks) (GH #158)
v.0.6.0 (November 25, 2011)¶
New Features¶
- Added melt function to pandas.core.reshape
- Added level parameter to group by level in Series and DataFrame descriptive statistics (PR313)
- Added head and tail methods to Series, analogous to to DataFrame (PR296)
- Added Series.isin function which checks if each value is contained in a passed sequence (GH289)
- Added float_format option to Series.to_string
- Added skip_footer (GH291) and converters (GH343) options to read_csv and read_table
- Added drop_duplicates and duplicated functions for removing duplicate DataFrame rows and checking for duplicate rows, respectively (GH319)
- Implemented operators ‘&’, ‘|’, ‘^’, ‘-‘ on DataFrame (GH347)
- Added Series.mad, mean absolute deviation
- Added QuarterEnd DateOffset (PR321)
- Added dot to DataFrame (GH65)
- Added orient option to Panel.from_dict (GH359, GH301)
- Added orient option to DataFrame.from_dict
- Added passing list of tuples or list of lists to DataFrame.from_records (GH357)
- Added multiple levels to groupby (GH103)
- Allow multiple columns in by argument of DataFrame.sort_index (GH92, PR362)
- Added fast get_value and put_value methods to DataFrame (GH360)
- Added cov instance methods to Series and DataFrame (GH194, PR362)
- Added kind='bar' option to DataFrame.plot (PR348)
- Added idxmin and idxmax to Series and DataFrame (PR286)
- Added read_clipboard function to parse DataFrame from clipboard (GH300)
- Added nunique function to Series for counting unique elements (GH297)
- Made DataFrame constructor use Series name if no columns passed (GH373)
- Support regular expressions in read_table/read_csv (GH364)
- Added DataFrame.to_html for writing DataFrame to HTML (PR387)
- Added support for MaskedArray data in DataFrame, masked values converted to NaN (PR396)
- Added DataFrame.boxplot function (GH368)
- Can pass extra args, kwds to DataFrame.apply (GH376)
- Implement DataFrame.join with vector on argument (GH312)
- Added legend boolean flag to DataFrame.plot (GH324)
- Can pass multiple levels to stack and unstack (GH370)
- Can pass multiple values columns to pivot_table (GH381)
- Use Series name in GroupBy for result index (GH363)
- Added raw option to DataFrame.apply for performance if only need ndarray (GH309)
- Added proper, tested weighted least squares to standard and panel OLS (GH303)
Performance Enhancements¶
- VBENCH Cythonized cache_readonly, resulting in substantial micro-performance enhancements throughout the codebase (GH361)
- VBENCH Special Cython matrix iterator for applying arbitrary reduction operations with 3-5x better performance than np.apply_along_axis (GH309)
- VBENCH Improved performance of MultiIndex.from_tuples
- VBENCH Special Cython matrix iterator for applying arbitrary reduction operations
- VBENCH + DOCUMENT Add raw option to DataFrame.apply for getting better performance when
- VBENCH Faster cythonized count by level in Series and DataFrame (GH341)
- VBENCH? Significant GroupBy performance enhancement with multiple keys with many “empty” combinations
- VBENCH New Cython vectorized function map_infer speeds up Series.apply and Series.map significantly when passed elementwise Python function, motivated by (PR355)
- VBENCH Significantly improved performance of Series.order, which also makes np.unique called on a Series faster (GH327)
- VBENCH Vastly improved performance of GroupBy on axes with a MultiIndex (GH299)
v.0.5.0 (October 24, 2011)¶
New Features¶
- Added DataFrame.align method with standard join options
- Added parse_dates option to read_csv and read_table methods to optionally try to parse dates in the index columns
- Added nrows, chunksize, and iterator arguments to read_csv and read_table. The last two return a new TextParser class capable of lazily iterating through chunks of a flat file (GH242)
- Added ability to join on multiple columns in DataFrame.join (GH214)
- Added private _get_duplicates function to Index for identifying duplicate values more easily (ENH5c)
- Added column attribute access to DataFrame.
- Added Python tab completion hook for DataFrame columns. (PR233, GH230)
- Implemented Series.describe for Series containing objects (PR241)
- Added inner join option to DataFrame.join when joining on key(s) (GH248)
- Implemented selecting DataFrame columns by passing a list to __getitem__ (GH253)
- Implemented & and | to intersect / union Index objects, respectively (GH261)
- Added pivot_table convenience function to pandas namespace (GH234)
- Implemented Panel.rename_axis function (GH243)
- DataFrame will show index level names in console output (PR334)
- Implemented Panel.take
- Added set_eng_float_format for alternate DataFrame floating point string formatting (ENH61)
- Added convenience set_index function for creating a DataFrame index from its existing columns
- Implemented groupby hierarchical index level name (GH223)
- Added support for different delimiters in DataFrame.to_csv (PR244)
- TODO: DOCS ABOUT TAKE METHODS
Performance Enhancements¶
- VBENCH Major performance improvements in file parsing functions read_csv and read_table
- VBENCH Added Cython function for converting tuples to ndarray very fast. Speeds up many MultiIndex-related operations
- VBENCH Refactored merging / joining code into a tidy class and disabled unnecessary computations in the float/object case, thus getting about 10% better performance (GH211)
- VBENCH Improved speed of DataFrame.xs on mixed-type DataFrame objects by about 5x, regression from 0.3.0 (GH215)
- VBENCH With new DataFrame.align method, speeding up binary operations between differently-indexed DataFrame objects by 10-25%.
- VBENCH Significantly sped up conversion of nested dict into DataFrame (GH212)
- VBENCH Significantly speed up DataFrame __repr__ and count on large mixed-type DataFrame objects
v.0.4.3 through v0.4.1 (September 25 - October 9, 2011)¶
New Features¶
- Added Python 3 support using 2to3 (PR200)
- Added name attribute to Series, now prints as part of Series.__repr__
- Added instance methods isnull and notnull to Series (PR209, GH203)
- Added Series.align method for aligning two series with choice of join method (ENH56)
- Added method get_level_values to MultiIndex (IS188)
- Set values in mixed-type DataFrame objects via .ix indexing attribute (GH135)
- Added new DataFrame methods get_dtype_counts and property dtypes (ENHdc)
- Added ignore_index option to DataFrame.append to stack DataFrames (ENH1b)
- read_csv tries to sniff delimiters using csv.Sniffer (PR146)
- read_csv can read multiple columns into a MultiIndex; DataFrame’s to_csv method writes out a corresponding MultiIndex (PR151)
- DataFrame.rename has a new copy parameter to rename a DataFrame in place (ENHed)
- Enable unstacking by name (PR142)
- Enable sortlevel to work by level (PR141)
Performance Enhancements¶
- Altered binary operations on differently-indexed SparseSeries objects to use the integer-based (dense) alignment logic which is faster with a larger number of blocks (GH205)
- Wrote faster Cython data alignment / merging routines resulting in substantial speed increases
- Improved performance of isnull and notnull, a regression from v0.3.0 (GH187)
- Refactored code related to DataFrame.join so that intermediate aligned copies of the data in each DataFrame argument do not need to be created. Substantial performance increases result (GH176)
- Substantially improved performance of generic Index.intersection and Index.union
- Implemented BlockManager.take resulting in significantly faster take performance on mixed-type DataFrame objects (GH104)
- Improved performance of Series.sort_index
- Significant groupby performance enhancement: removed unnecessary integrity checks in DataFrame internals that were slowing down slicing operations to retrieve groups
- Optimized _ensure_index function resulting in performance savings in type-checking Index objects
- Wrote fast time series merging / joining methods in Cython. Will be integrated later into DataFrame.join and related functions