Cookbook¶
This is a repository for short and sweet examples and links for useful pandas recipes. We encourage users to add to this documentation.
Adding interesting links and/or inline examples to this section is a great First Pull Request.
Simplified, condensed, new-user friendly, in-line examples have been inserted where possible to augment the Stack-Overflow and GitHub links. Many of the links contain expanded information, above what the in-line examples offer.
Pandas (pd) and Numpy (np) are the only two abbreviated imported modules. The rest are kept explicitly imported for newer users.
These examples are written for python 3.4. Minor tweaks might be necessary for earlier python versions.
Idioms¶
These are some neat pandas idioms
if-then/if-then-else on one column, and assignment to another one or more columns:
In [1]: df = pd.DataFrame(
...: {'AAA' : [4,5,6,7], 'BBB' : [10,20,30,40],'CCC' : [100,50,-30,-50]}); df
...:
Out[1]:
AAA BBB CCC
0 4 10 100
1 5 20 50
2 6 30 -30
3 7 40 -50
if-then...¶
An if-then on one column
In [2]: df.ix[df.AAA >= 5,'BBB'] = -1; df
Out[2]:
AAA BBB CCC
0 4 10 100
1 5 -1 50
2 6 -1 -30
3 7 -1 -50
An if-then with assignment to 2 columns:
In [3]: df.ix[df.AAA >= 5,['BBB','CCC']] = 555; df
Out[3]:
AAA BBB CCC
0 4 10 100
1 5 555 555
2 6 555 555
3 7 555 555
Add another line with different logic, to do the -else
In [4]: df.ix[df.AAA < 5,['BBB','CCC']] = 2000; df
Out[4]:
AAA BBB CCC
0 4 2000 2000
1 5 555 555
2 6 555 555
3 7 555 555
Or use pandas where after you’ve set up a mask
In [5]: df_mask = pd.DataFrame({'AAA' : [True] * 4, 'BBB' : [False] * 4,'CCC' : [True,False] * 2})
In [6]: df.where(df_mask,-1000)
Out[6]:
AAA BBB CCC
0 4 -1000 2000
1 5 -1000 -1000
2 6 -1000 555
3 7 -1000 -1000
if-then-else using numpy’s where()
In [7]: df = pd.DataFrame(
...: {'AAA' : [4,5,6,7], 'BBB' : [10,20,30,40],'CCC' : [100,50,-30,-50]}); df
...:
Out[7]:
AAA BBB CCC
0 4 10 100
1 5 20 50
2 6 30 -30
3 7 40 -50
In [8]: df['logic'] = np.where(df['AAA'] > 5,'high','low'); df
Out[8]:
AAA BBB CCC logic
0 4 10 100 low
1 5 20 50 low
2 6 30 -30 high
3 7 40 -50 high
Splitting¶
Split a frame with a boolean criterion
In [9]: df = pd.DataFrame(
...: {'AAA' : [4,5,6,7], 'BBB' : [10,20,30,40],'CCC' : [100,50,-30,-50]}); df
...:
Out[9]:
AAA BBB CCC
0 4 10 100
1 5 20 50
2 6 30 -30
3 7 40 -50
In [10]: dflow = df[df.AAA <= 5]
In [11]: dfhigh = df[df.AAA > 5]
In [12]: dflow; dfhigh
Out[12]:
AAA BBB CCC
2 6 30 -30
3 7 40 -50
Building Criteria¶
Select with multi-column criteria
In [13]: df = pd.DataFrame(
....: {'AAA' : [4,5,6,7], 'BBB' : [10,20,30,40],'CCC' : [100,50,-30,-50]}); df
....:
Out[13]:
AAA BBB CCC
0 4 10 100
1 5 20 50
2 6 30 -30
3 7 40 -50
...and (without assignment returns a Series)
In [14]: newseries = df.loc[(df['BBB'] < 25) & (df['CCC'] >= -40), 'AAA']; newseries
Out[14]:
0 4
1 5
Name: AAA, dtype: int64
...or (without assignment returns a Series)
In [15]: newseries = df.loc[(df['BBB'] > 25) | (df['CCC'] >= -40), 'AAA']; newseries;
...or (with assignment modifies the DataFrame.)
In [16]: df.loc[(df['BBB'] > 25) | (df['CCC'] >= 75), 'AAA'] = 0.1; df
Out[16]:
AAA BBB CCC
0 0.1 10 100
1 5.0 20 50
2 0.1 30 -30
3 0.1 40 -50
Select rows with data closest to certain value using argsort
In [17]: df = pd.DataFrame(
....: {'AAA' : [4,5,6,7], 'BBB' : [10,20,30,40],'CCC' : [100,50,-30,-50]}); df
....:
Out[17]:
AAA BBB CCC
0 4 10 100
1 5 20 50
2 6 30 -30
3 7 40 -50
In [18]: aValue = 43.0
In [19]: df.ix[(df.CCC-aValue).abs().argsort()]
Out[19]:
AAA BBB CCC
1 5 20 50
0 4 10 100
2 6 30 -30
3 7 40 -50
Dynamically reduce a list of criteria using a binary operators
In [20]: df = pd.DataFrame(
....: {'AAA' : [4,5,6,7], 'BBB' : [10,20,30,40],'CCC' : [100,50,-30,-50]}); df
....:
Out[20]:
AAA BBB CCC
0 4 10 100
1 5 20 50
2 6 30 -30
3 7 40 -50
In [21]: Crit1 = df.AAA <= 5.5
In [22]: Crit2 = df.BBB == 10.0
In [23]: Crit3 = df.CCC > -40.0
One could hard code:
In [24]: AllCrit = Crit1 & Crit2 & Crit3
...Or it can be done with a list of dynamically built criteria
In [25]: CritList = [Crit1,Crit2,Crit3]
In [26]: AllCrit = functools.reduce(lambda x,y: x & y, CritList)
In [27]: df[AllCrit]
Out[27]:
AAA BBB CCC
0 4 10 100
Selection¶
DataFrames¶
The indexing docs.
Using both row labels and value conditionals
In [28]: df = pd.DataFrame(
....: {'AAA' : [4,5,6,7], 'BBB' : [10,20,30,40],'CCC' : [100,50,-30,-50]}); df
....:
Out[28]:
AAA BBB CCC
0 4 10 100
1 5 20 50
2 6 30 -30
3 7 40 -50
In [29]: df[(df.AAA <= 6) & (df.index.isin([0,2,4]))]
Out[29]:
AAA BBB CCC
0 4 10 100
2 6 30 -30
Use loc for label-oriented slicing and iloc positional slicing
In [30]: data = {'AAA' : [4,5,6,7], 'BBB' : [10,20,30,40],'CCC' : [100,50,-30,-50]}
In [31]: df = pd.DataFrame(data=data,index=['foo','bar','boo','kar']); df
Out[31]:
AAA BBB CCC
foo 4 10 100
bar 5 20 50
boo 6 30 -30
kar 7 40 -50
There are 2 explicit slicing methods, with a third general case
- Positional-oriented (Python slicing style : exclusive of end)
- Label-oriented (Non-Python slicing style : inclusive of end)
- General (Either slicing style : depends on if the slice contains labels or positions)
In [32]: df.loc['bar':'kar'] #Label
Out[32]:
AAA BBB CCC
bar 5 20 50
boo 6 30 -30
kar 7 40 -50
#Generic
In [33]: df.ix[0:3] #Same as .iloc[0:3]
Out[33]:
AAA BBB CCC
foo 4 10 100
bar 5 20 50
boo 6 30 -30
In [34]: df.ix['bar':'kar'] #Same as .loc['bar':'kar']
Out[34]:
AAA BBB CCC
bar 5 20 50
boo 6 30 -30
kar 7 40 -50
Ambiguity arises when an index consists of integers with a non-zero start or non-unit increment.
In [35]: df2 = pd.DataFrame(data=data,index=[1,2,3,4]); #Note index starts at 1.
In [36]: df2.iloc[1:3] #Position-oriented
Out[36]:
AAA BBB CCC
2 5 20 50
3 6 30 -30
In [37]: df2.loc[1:3] #Label-oriented
Out[37]:
AAA BBB CCC
1 4 10 100
2 5 20 50
3 6 30 -30
In [38]: df2.ix[1:3] #General, will mimic loc (label-oriented)
Out[38]:
AAA BBB CCC
1 4 10 100
2 5 20 50
3 6 30 -30
In [39]: df2.ix[0:3] #General, will mimic iloc (position-oriented), as loc[0:3] would raise a KeyError
Out[39]:
AAA BBB CCC
1 4 10 100
2 5 20 50
3 6 30 -30
Using inverse operator (~) to take the complement of a mask
In [40]: df = pd.DataFrame(
....: {'AAA' : [4,5,6,7], 'BBB' : [10,20,30,40], 'CCC' : [100,50,-30,-50]}); df
....:
Out[40]:
AAA BBB CCC
0 4 10 100
1 5 20 50
2 6 30 -30
3 7 40 -50
In [41]: df[~((df.AAA <= 6) & (df.index.isin([0,2,4])))]
Out[41]:
AAA BBB CCC
1 5 20 50
3 7 40 -50
Panels¶
In [42]: rng = pd.date_range('1/1/2013',periods=100,freq='D')
In [43]: data = np.random.randn(100, 4)
In [44]: cols = ['A','B','C','D']
In [45]: df1, df2, df3 = pd.DataFrame(data, rng, cols), pd.DataFrame(data, rng, cols), pd.DataFrame(data, rng, cols)
In [46]: pf = pd.Panel({'df1':df1,'df2':df2,'df3':df3});pf
Out[46]:
<class 'pandas.core.panel.Panel'>
Dimensions: 3 (items) x 100 (major_axis) x 4 (minor_axis)
Items axis: df1 to df3
Major_axis axis: 2013-01-01 00:00:00 to 2013-04-10 00:00:00
Minor_axis axis: A to D
#Assignment using Transpose (pandas < 0.15)
In [47]: pf = pf.transpose(2,0,1)
In [48]: pf['E'] = pd.DataFrame(data, rng, cols)
In [49]: pf = pf.transpose(1,2,0);pf
Out[49]:
<class 'pandas.core.panel.Panel'>
Dimensions: 3 (items) x 100 (major_axis) x 5 (minor_axis)
Items axis: df1 to df3
Major_axis axis: 2013-01-01 00:00:00 to 2013-04-10 00:00:00
Minor_axis axis: A to E
#Direct assignment (pandas > 0.15)
In [50]: pf.loc[:,:,'F'] = pd.DataFrame(data, rng, cols);pf
Out[50]:
<class 'pandas.core.panel.Panel'>
Dimensions: 3 (items) x 100 (major_axis) x 6 (minor_axis)
Items axis: df1 to df3
Major_axis axis: 2013-01-01 00:00:00 to 2013-04-10 00:00:00
Minor_axis axis: A to F
Mask a panel by using np.where and then reconstructing the panel with the new masked values
New Columns¶
Efficiently and dynamically creating new columns using applymap
In [51]: df = pd.DataFrame(
....: {'AAA' : [1,2,1,3], 'BBB' : [1,1,2,2], 'CCC' : [2,1,3,1]}); df
....:
Out[51]:
AAA BBB CCC
0 1 1 2
1 2 1 1
2 1 2 3
3 3 2 1
In [52]: source_cols = df.columns # or some subset would work too.
In [53]: new_cols = [str(x) + "_cat" for x in source_cols]
In [54]: categories = {1 : 'Alpha', 2 : 'Beta', 3 : 'Charlie' }
In [55]: df[new_cols] = df[source_cols].applymap(categories.get);df
Out[55]:
AAA BBB CCC AAA_cat BBB_cat CCC_cat
0 1 1 2 Alpha Alpha Beta
1 2 1 1 Beta Alpha Alpha
2 1 2 3 Alpha Beta Charlie
3 3 2 1 Charlie Beta Alpha
Keep other columns when using min() with groupby
In [56]: df = pd.DataFrame(
....: {'AAA' : [1,1,1,2,2,2,3,3], 'BBB' : [2,1,3,4,5,1,2,3]}); df
....:
Out[56]:
AAA BBB
0 1 2
1 1 1
2 1 3
3 2 4
4 2 5
5 2 1
6 3 2
7 3 3
Method 1 : idxmin() to get the index of the mins
In [57]: df.loc[df.groupby("AAA")["BBB"].idxmin()]
Out[57]:
AAA BBB
1 1 1
5 2 1
6 3 2
Method 2 : sort then take first of each
In [58]: df.sort_values(by="BBB").groupby("AAA", as_index=False).first()
Out[58]:
AAA BBB
0 1 1
1 2 1
2 3 2
Notice the same results, with the exception of the index.
MultiIndexing¶
The multindexing docs.
Creating a multi-index from a labeled frame
In [59]: df = pd.DataFrame({'row' : [0,1,2],
....: 'One_X' : [1.1,1.1,1.1],
....: 'One_Y' : [1.2,1.2,1.2],
....: 'Two_X' : [1.11,1.11,1.11],
....: 'Two_Y' : [1.22,1.22,1.22]}); df
....:
Out[59]:
One_X One_Y Two_X Two_Y row
0 1.1 1.2 1.11 1.22 0
1 1.1 1.2 1.11 1.22 1
2 1.1 1.2 1.11 1.22 2
# As Labelled Index
In [60]: df = df.set_index('row');df
Out[60]:
One_X One_Y Two_X Two_Y
row
0 1.1 1.2 1.11 1.22
1 1.1 1.2 1.11 1.22
2 1.1 1.2 1.11 1.22
# With Hierarchical Columns
In [61]: df.columns = pd.MultiIndex.from_tuples([tuple(c.split('_')) for c in df.columns]);df
Out[61]:
One Two
X Y X Y
row
0 1.1 1.2 1.11 1.22
1 1.1 1.2 1.11 1.22
2 1.1 1.2 1.11 1.22
# Now stack & Reset
In [62]: df = df.stack(0).reset_index(1);df
Out[62]:
level_1 X Y
row
0 One 1.10 1.20
0 Two 1.11 1.22
1 One 1.10 1.20
1 Two 1.11 1.22
2 One 1.10 1.20
2 Two 1.11 1.22
# And fix the labels (Notice the label 'level_1' got added automatically)
In [63]: df.columns = ['Sample','All_X','All_Y'];df
Out[63]:
Sample All_X All_Y
row
0 One 1.10 1.20
0 Two 1.11 1.22
1 One 1.10 1.20
1 Two 1.11 1.22
2 One 1.10 1.20
2 Two 1.11 1.22
Arithmetic¶
Performing arithmetic with a multi-index that needs broadcasting
In [64]: cols = pd.MultiIndex.from_tuples([ (x,y) for x in ['A','B','C'] for y in ['O','I']])
In [65]: df = pd.DataFrame(np.random.randn(2,6),index=['n','m'],columns=cols); df
Out[65]:
A B C
O I O I O I
n 1.920906 -0.388231 -2.314394 0.665508 0.402562 0.399555
m -1.765956 0.850423 0.388054 0.992312 0.744086 -0.739776
In [66]: df = df.div(df['C'],level=1); df
Out[66]:
A B C
O I O I O I
n 4.771702 -0.971660 -5.749162 1.665625 1.0 1.0
m -2.373321 -1.149568 0.521518 -1.341367 1.0 1.0
Slicing¶
In [67]: coords = [('AA','one'),('AA','six'),('BB','one'),('BB','two'),('BB','six')]
In [68]: index = pd.MultiIndex.from_tuples(coords)
In [69]: df = pd.DataFrame([11,22,33,44,55],index,['MyData']); df
Out[69]:
MyData
AA one 11
six 22
BB one 33
two 44
six 55
To take the cross section of the 1st level and 1st axis the index:
In [70]: df.xs('BB',level=0,axis=0) #Note : level and axis are optional, and default to zero
Out[70]:
MyData
one 33
two 44
six 55
...and now the 2nd level of the 1st axis.
In [71]: df.xs('six',level=1,axis=0)
Out[71]:
MyData
AA 22
BB 55
Slicing a multi-index with xs, method #2
In [72]: index = list(itertools.product(['Ada','Quinn','Violet'],['Comp','Math','Sci']))
In [73]: headr = list(itertools.product(['Exams','Labs'],['I','II']))
In [74]: indx = pd.MultiIndex.from_tuples(index,names=['Student','Course'])
In [75]: cols = pd.MultiIndex.from_tuples(headr) #Notice these are un-named
In [76]: data = [[70+x+y+(x*y)%3 for x in range(4)] for y in range(9)]
In [77]: df = pd.DataFrame(data,indx,cols); df
Out[77]:
Exams Labs
I II I II
Student Course
Ada Comp 70 71 72 73
Math 71 73 75 74
Sci 72 75 75 75
Quinn Comp 73 74 75 76
Math 74 76 78 77
Sci 75 78 78 78
Violet Comp 76 77 78 79
Math 77 79 81 80
Sci 78 81 81 81
In [78]: All = slice(None)
In [79]: df.loc['Violet']
Out[79]:
Exams Labs
I II I II
Course
Comp 76 77 78 79
Math 77 79 81 80
Sci 78 81 81 81
In [80]: df.loc[(All,'Math'),All]
Out[80]:
Exams Labs
I II I II
Student Course
Ada Math 71 73 75 74
Quinn Math 74 76 78 77
Violet Math 77 79 81 80
In [81]: df.loc[(slice('Ada','Quinn'),'Math'),All]
Out[81]:
Exams Labs
I II I II
Student Course
Ada Math 71 73 75 74
Quinn Math 74 76 78 77
In [82]: df.loc[(All,'Math'),('Exams')]
Out[82]:
I II
Student Course
Ada Math 71 73
Quinn Math 74 76
Violet Math 77 79
In [83]: df.loc[(All,'Math'),(All,'II')]
Out[83]:
Exams Labs
II II
Student Course
Ada Math 73 74
Quinn Math 76 77
Violet Math 79 80
Sorting¶
Sort by specific column or an ordered list of columns, with a multi-index
In [84]: df.sort_values(by=('Labs', 'II'), ascending=False)
Out[84]:
Exams Labs
I II I II
Student Course
Violet Sci 78 81 81 81
Math 77 79 81 80
Comp 76 77 78 79
Quinn Sci 75 78 78 78
Math 74 76 78 77
Comp 73 74 75 76
Ada Sci 72 75 75 75
Math 71 73 75 74
Comp 70 71 72 73
Missing Data¶
The missing data docs.
Fill forward a reversed timeseries
In [85]: df = pd.DataFrame(np.random.randn(6,1), index=pd.date_range('2013-08-01', periods=6, freq='B'), columns=list('A'))
In [86]: df.ix[3,'A'] = np.nan
In [87]: df
Out[87]:
A
2013-08-01 -1.054874
2013-08-02 -0.179642
2013-08-05 0.639589
2013-08-06 NaN
2013-08-07 1.906684
2013-08-08 0.104050
In [88]: df.reindex(df.index[::-1]).ffill()
Out[88]:
A
2013-08-08 0.104050
2013-08-07 1.906684
2013-08-06 1.906684
2013-08-05 0.639589
2013-08-02 -0.179642
2013-08-01 -1.054874
Replace¶
Grouping¶
The grouping docs.
Unlike agg, apply’s callable is passed a sub-DataFrame which gives you access to all the columns
In [89]: df = pd.DataFrame({'animal': 'cat dog cat fish dog cat cat'.split(),
....: 'size': list('SSMMMLL'),
....: 'weight': [8, 10, 11, 1, 20, 12, 12],
....: 'adult' : [False] * 5 + [True] * 2}); df
....:
Out[89]:
adult animal size weight
0 False cat S 8
1 False dog S 10
2 False cat M 11
3 False fish M 1
4 False dog M 20
5 True cat L 12
6 True cat L 12
#List the size of the animals with the highest weight.
In [90]: df.groupby('animal').apply(lambda subf: subf['size'][subf['weight'].idxmax()])
Out[90]:
animal
cat L
dog M
fish M
dtype: object
In [91]: gb = df.groupby(['animal'])
In [92]: gb.get_group('cat')
Out[92]:
adult animal size weight
0 False cat S 8
2 False cat M 11
5 True cat L 12
6 True cat L 12
Apply to different items in a group
In [93]: def GrowUp(x):
....: avg_weight = sum(x[x['size'] == 'S'].weight * 1.5)
....: avg_weight += sum(x[x['size'] == 'M'].weight * 1.25)
....: avg_weight += sum(x[x['size'] == 'L'].weight)
....: avg_weight /= len(x)
....: return pd.Series(['L',avg_weight,True], index=['size', 'weight', 'adult'])
....:
In [94]: expected_df = gb.apply(GrowUp)
In [95]: expected_df
Out[95]:
size weight adult
animal
cat L 12.4375 True
dog L 20.0000 True
fish L 1.2500 True
In [96]: S = pd.Series([i / 100.0 for i in range(1,11)])
In [97]: def CumRet(x,y):
....: return x * (1 + y)
....:
In [98]: def Red(x):
....: return functools.reduce(CumRet,x,1.0)
....:
In [99]: S.expanding().apply(Red)
Out[99]:
0 1.010000
1 1.030200
2 1.061106
3 1.103550
4 1.158728
5 1.228251
6 1.314229
7 1.419367
8 1.547110
9 1.701821
dtype: float64
Replacing some values with mean of the rest of a group
In [100]: df = pd.DataFrame({'A' : [1, 1, 2, 2], 'B' : [1, -1, 1, 2]})
In [101]: gb = df.groupby('A')
In [102]: def replace(g):
.....: mask = g < 0
.....: g.loc[mask] = g[~mask].mean()
.....: return g
.....:
In [103]: gb.transform(replace)
Out[103]:
B
0 1.0
1 1.0
2 1.0
3 2.0
Sort groups by aggregated data
In [104]: df = pd.DataFrame({'code': ['foo', 'bar', 'baz'] * 2,
.....: 'data': [0.16, -0.21, 0.33, 0.45, -0.59, 0.62],
.....: 'flag': [False, True] * 3})
.....:
In [105]: code_groups = df.groupby('code')
In [106]: agg_n_sort_order = code_groups[['data']].transform(sum).sort_values(by='data')
In [107]: sorted_df = df.ix[agg_n_sort_order.index]
In [108]: sorted_df
Out[108]:
code data flag
1 bar -0.21 True
4 bar -0.59 False
0 foo 0.16 False
3 foo 0.45 True
2 baz 0.33 False
5 baz 0.62 True
Create multiple aggregated columns
In [109]: rng = pd.date_range(start="2014-10-07",periods=10,freq='2min')
In [110]: ts = pd.Series(data = list(range(10)), index = rng)
In [111]: def MyCust(x):
.....: if len(x) > 2:
.....: return x[1] * 1.234
.....: return pd.NaT
.....:
In [112]: mhc = {'Mean' : np.mean, 'Max' : np.max, 'Custom' : MyCust}
In [113]: ts.resample("5min").apply(mhc)
Out[113]:
Max Custom Mean
2014-10-07 00:00:00 2 1.234 1.0
2014-10-07 00:05:00 4 NaT 3.5
2014-10-07 00:10:00 7 7.404 6.0
2014-10-07 00:15:00 9 NaT 8.5
In [114]: ts
Out[114]:
2014-10-07 00:00:00 0
2014-10-07 00:02:00 1
2014-10-07 00:04:00 2
2014-10-07 00:06:00 3
2014-10-07 00:08:00 4
2014-10-07 00:10:00 5
2014-10-07 00:12:00 6
2014-10-07 00:14:00 7
2014-10-07 00:16:00 8
2014-10-07 00:18:00 9
Freq: 2T, dtype: int64
Create a value counts column and reassign back to the DataFrame
In [115]: df = pd.DataFrame({'Color': 'Red Red Red Blue'.split(),
.....: 'Value': [100, 150, 50, 50]}); df
.....:
Out[115]:
Color Value
0 Red 100
1 Red 150
2 Red 50
3 Blue 50
In [116]: df['Counts'] = df.groupby(['Color']).transform(len)
In [117]: df
Out[117]:
Color Value Counts
0 Red 100 3
1 Red 150 3
2 Red 50 3
3 Blue 50 1
Shift groups of the values in a column based on the index
In [118]: df = pd.DataFrame(
.....: {u'line_race': [10, 10, 8, 10, 10, 8],
.....: u'beyer': [99, 102, 103, 103, 88, 100]},
.....: index=[u'Last Gunfighter', u'Last Gunfighter', u'Last Gunfighter',
.....: u'Paynter', u'Paynter', u'Paynter']); df
.....:
Out[118]:
beyer line_race
Last Gunfighter 99 10
Last Gunfighter 102 10
Last Gunfighter 103 8
Paynter 103 10
Paynter 88 10
Paynter 100 8
In [119]: df['beyer_shifted'] = df.groupby(level=0)['beyer'].shift(1)
In [120]: df
Out[120]:
beyer line_race beyer_shifted
Last Gunfighter 99 10 NaN
Last Gunfighter 102 10 99.0
Last Gunfighter 103 8 102.0
Paynter 103 10 NaN
Paynter 88 10 103.0
Paynter 100 8 88.0
Select row with maximum value from each group
In [121]: df = pd.DataFrame({'host':['other','other','that','this','this'],
.....: 'service':['mail','web','mail','mail','web'],
.....: 'no':[1, 2, 1, 2, 1]}).set_index(['host', 'service'])
.....:
In [122]: mask = df.groupby(level=0).agg('idxmax')
In [123]: df_count = df.loc[mask['no']].reset_index()
In [124]: df_count
Out[124]:
host service no
0 other web 2
1 that mail 1
2 this mail 2
Grouping like Python’s itertools.groupby
In [125]: df = pd.DataFrame([0, 1, 0, 1, 1, 1, 0, 1, 1], columns=['A'])
In [126]: df.A.groupby((df.A != df.A.shift()).cumsum()).groups
Out[126]:
{1: Int64Index([0], dtype='int64'),
2: Int64Index([1], dtype='int64'),
3: Int64Index([2], dtype='int64'),
4: Int64Index([3, 4, 5], dtype='int64'),
5: Int64Index([6], dtype='int64'),
6: Int64Index([7, 8], dtype='int64')}
In [127]: df.A.groupby((df.A != df.A.shift()).cumsum()).cumsum()
Out[127]:
0 0
1 1
2 0
3 1
4 2
5 3
6 0
7 1
8 2
Name: A, dtype: int64
Splitting¶
Create a list of dataframes, split using a delineation based on logic included in rows.
In [128]: df = pd.DataFrame(data={'Case' : ['A','A','A','B','A','A','B','A','A'],
.....: 'Data' : np.random.randn(9)})
.....:
In [129]: dfs = list(zip(*df.groupby((1*(df['Case']=='B')).cumsum().rolling(window=3,min_periods=1).median())))[-1]
In [130]: dfs[0]
Out[130]:
Case Data
0 A 0.174068
1 A -0.439461
2 A -0.741343
3 B -0.079673
In [131]: dfs[1]
Out[131]:
Case Data
4 A -0.922875
5 A 0.303638
6 B -0.917368
In [132]: dfs[2]
Out[132]:
Case Data
7 A -1.624062
8 A -0.758514
Pivot¶
The Pivot docs.
In [133]: df = pd.DataFrame(data={'Province' : ['ON','QC','BC','AL','AL','MN','ON'],
.....: 'City' : ['Toronto','Montreal','Vancouver','Calgary','Edmonton','Winnipeg','Windsor'],
.....: 'Sales' : [13,6,16,8,4,3,1]})
.....:
In [134]: table = pd.pivot_table(df,values=['Sales'],index=['Province'],columns=['City'],aggfunc=np.sum,margins=True)
In [135]: table.stack('City')
Out[135]:
Sales
Province City
AL All 12.0
Calgary 8.0
Edmonton 4.0
BC All 16.0
Vancouver 16.0
MN All 3.0
Winnipeg 3.0
... ...
All Calgary 8.0
Edmonton 4.0
Montreal 6.0
Toronto 13.0
Vancouver 16.0
Windsor 1.0
Winnipeg 3.0
[20 rows x 1 columns]
Frequency table like plyr in R
In [136]: grades = [48,99,75,80,42,80,72,68,36,78]
In [137]: df = pd.DataFrame( {'ID': ["x%d" % r for r in range(10)],
.....: 'Gender' : ['F', 'M', 'F', 'M', 'F', 'M', 'F', 'M', 'M', 'M'],
.....: 'ExamYear': ['2007','2007','2007','2008','2008','2008','2008','2009','2009','2009'],
.....: 'Class': ['algebra', 'stats', 'bio', 'algebra', 'algebra', 'stats', 'stats', 'algebra', 'bio', 'bio'],
.....: 'Participated': ['yes','yes','yes','yes','no','yes','yes','yes','yes','yes'],
.....: 'Passed': ['yes' if x > 50 else 'no' for x in grades],
.....: 'Employed': [True,True,True,False,False,False,False,True,True,False],
.....: 'Grade': grades})
.....:
In [138]: df.groupby('ExamYear').agg({'Participated': lambda x: x.value_counts()['yes'],
.....: 'Passed': lambda x: sum(x == 'yes'),
.....: 'Employed' : lambda x : sum(x),
.....: 'Grade' : lambda x : sum(x) / len(x)})
.....:
Out[138]:
Grade Employed Participated Passed
ExamYear
2007 74 3 3 2
2008 68 0 3 3
2009 60 2 3 2
Plot pandas DataFrame with year over year data
To create year and month crosstabulation:
In [139]: df = pd.DataFrame({'value': np.random.randn(36)},
.....: index=pd.date_range('2011-01-01', freq='M', periods=36))
.....:
In [140]: pd.pivot_table(df, index=df.index.month, columns=df.index.year,
.....: values='value', aggfunc='sum')
.....:
Out[140]:
2011 2012 2013
1 -0.560859 0.120930 0.516870
2 -0.589005 -0.210518 0.343125
3 -1.070678 -0.931184 2.137827
4 -1.681101 0.240647 0.452429
5 0.403776 -0.027462 0.483103
6 0.609862 0.033113 0.061495
7 0.387936 -0.658418 0.240767
8 1.815066 0.324102 0.782413
9 0.705200 -1.403048 0.628462
10 -0.668049 -0.581967 -0.880627
11 0.242501 -1.233862 0.777575
12 0.313421 -3.520876 -0.779367
Apply¶
Rolling Apply to Organize - Turning embedded lists into a multi-index frame
In [141]: df = pd.DataFrame(data={'A' : [[2,4,8,16],[100,200],[10,20,30]], 'B' : [['a','b','c'],['jj','kk'],['ccc']]},index=['I','II','III'])
In [142]: def SeriesFromSubList(aList):
.....: return pd.Series(aList)
.....:
In [143]: df_orgz = pd.concat(dict([ (ind,row.apply(SeriesFromSubList)) for ind,row in df.iterrows() ]))
Rolling Apply with a DataFrame returning a Series
Rolling Apply to multiple columns where function calculates a Series before a Scalar from the Series is returned
In [144]: df = pd.DataFrame(data=np.random.randn(2000,2)/10000,
.....: index=pd.date_range('2001-01-01',periods=2000),
.....: columns=['A','B']); df
.....:
Out[144]:
A B
2001-01-01 0.000032 -0.000004
2001-01-02 -0.000001 0.000207
2001-01-03 0.000120 -0.000220
2001-01-04 -0.000083 -0.000165
2001-01-05 -0.000047 0.000156
2001-01-06 0.000027 0.000104
2001-01-07 0.000041 -0.000101
... ... ...
2006-06-17 -0.000034 0.000034
2006-06-18 0.000002 0.000166
2006-06-19 0.000023 -0.000081
2006-06-20 -0.000061 0.000012
2006-06-21 -0.000111 0.000027
2006-06-22 -0.000061 -0.000009
2006-06-23 0.000074 -0.000138
[2000 rows x 2 columns]
In [145]: def gm(aDF,Const):
.....: v = ((((aDF.A+aDF.B)+1).cumprod())-1)*Const
.....: return (aDF.index[0],v.iloc[-1])
.....:
In [146]: S = pd.Series(dict([ gm(df.iloc[i:min(i+51,len(df)-1)],5) for i in range(len(df)-50) ])); S
Out[146]:
2001-01-01 -0.001373
2001-01-02 -0.001705
2001-01-03 -0.002885
2001-01-04 -0.002987
2001-01-05 -0.002384
2001-01-06 -0.004700
2001-01-07 -0.005500
...
2006-04-28 -0.002682
2006-04-29 -0.002436
2006-04-30 -0.002602
2006-05-01 -0.001785
2006-05-02 -0.001799
2006-05-03 -0.000605
2006-05-04 -0.000541
dtype: float64
Rolling apply with a DataFrame returning a Scalar
Rolling Apply to multiple columns where function returns a Scalar (Volume Weighted Average Price)
In [147]: rng = pd.date_range(start = '2014-01-01',periods = 100)
In [148]: df = pd.DataFrame({'Open' : np.random.randn(len(rng)),
.....: 'Close' : np.random.randn(len(rng)),
.....: 'Volume' : np.random.randint(100,2000,len(rng))}, index=rng); df
.....:
Out[148]:
Close Open Volume
2014-01-01 -0.653039 0.011174 1581
2014-01-02 1.314205 0.214258 1707
2014-01-03 -0.341915 -1.046922 1768
2014-01-04 -1.303586 -0.752902 836
2014-01-05 0.396288 -0.410793 694
2014-01-06 -0.548006 0.648401 796
2014-01-07 0.481380 0.737320 265
... ... ... ...
2014-04-04 -2.548128 0.120378 564
2014-04-05 0.223346 0.231661 1908
2014-04-06 1.228841 0.952664 1090
2014-04-07 0.552784 -0.176090 1813
2014-04-08 -0.795389 1.781318 1103
2014-04-09 -0.018815 -0.753493 1456
2014-04-10 1.138197 -1.047997 1193
[100 rows x 3 columns]
In [149]: def vwap(bars): return ((bars.Close*bars.Volume).sum()/bars.Volume.sum())
In [150]: window = 5
In [151]: s = pd.concat([ (pd.Series(vwap(df.iloc[i:i+window]), index=[df.index[i+window]])) for i in range(len(df)-window) ]);
In [152]: s.round(2)
Out[152]:
2014-01-06 -0.03
2014-01-07 0.07
2014-01-08 -0.40
2014-01-09 -0.81
2014-01-10 -0.63
2014-01-11 -0.86
2014-01-12 -0.36
...
2014-04-04 -1.27
2014-04-05 -1.36
2014-04-06 -0.73
2014-04-07 0.04
2014-04-08 0.21
2014-04-09 0.07
2014-04-10 0.25
dtype: float64
Timeseries¶
Constructing a datetime range that excludes weekends and includes only certain times
Aggregation and plotting time series
Turn a matrix with hours in columns and days in rows into a continuous row sequence in the form of a time series. How to rearrange a python pandas DataFrame?
Dealing with duplicates when reindexing a timeseries to a specified frequency
Calculate the first day of the month for each entry in a DatetimeIndex
In [153]: dates = pd.date_range('2000-01-01', periods=5)
In [154]: dates.to_period(freq='M').to_timestamp()
Out[154]:
DatetimeIndex(['2000-01-01', '2000-01-01', '2000-01-01', '2000-01-01',
'2000-01-01'],
dtype='datetime64[ns]', freq=None)
Merge¶
The Concat docs. The Join docs.
Append two dataframes with overlapping index (emulate R rbind)
In [155]: rng = pd.date_range('2000-01-01', periods=6)
In [156]: df1 = pd.DataFrame(np.random.randn(6, 3), index=rng, columns=['A', 'B', 'C'])
In [157]: df2 = df1.copy()
ignore_index is needed in pandas < v0.13, and depending on df construction
In [158]: df = df1.append(df2,ignore_index=True); df
Out[158]:
A B C
0 -0.480676 -1.305282 -0.212846
1 1.979901 0.363112 -0.275732
2 -1.433852 0.580237 -0.013672
3 1.776623 -0.803467 0.521517
4 -0.302508 -0.442948 -0.395768
5 -0.249024 -0.031510 2.413751
6 -0.480676 -1.305282 -0.212846
7 1.979901 0.363112 -0.275732
8 -1.433852 0.580237 -0.013672
9 1.776623 -0.803467 0.521517
10 -0.302508 -0.442948 -0.395768
11 -0.249024 -0.031510 2.413751
In [159]: df = pd.DataFrame(data={'Area' : ['A'] * 5 + ['C'] * 2,
.....: 'Bins' : [110] * 2 + [160] * 3 + [40] * 2,
.....: 'Test_0' : [0, 1, 0, 1, 2, 0, 1],
.....: 'Data' : np.random.randn(7)});df
.....:
Out[159]:
Area Bins Data Test_0
0 A 110 -0.378914 0
1 A 110 -1.032527 1
2 A 160 -1.402816 0
3 A 160 0.715333 1
4 A 160 -0.091438 2
5 C 40 1.608418 0
6 C 40 0.753207 1
In [160]: df['Test_1'] = df['Test_0'] - 1
In [161]: pd.merge(df, df, left_on=['Bins', 'Area','Test_0'], right_on=['Bins', 'Area','Test_1'],suffixes=('_L','_R'))
Out[161]:
Area Bins Data_L Test_0_L Test_1_L Data_R Test_0_R Test_1_R
0 A 110 -0.378914 0 -1 -1.032527 1 0
1 A 160 -1.402816 0 -1 0.715333 1 0
2 A 160 0.715333 1 0 -0.091438 2 1
3 C 40 1.608418 0 -1 0.753207 1 0
Plotting¶
The Plotting docs.
Setting x-axis major and minor labels
Plotting multiple charts in an ipython notebook
Annotate a time-series plot #2
Generate Embedded plots in excel files using Pandas, Vincent and xlsxwriter
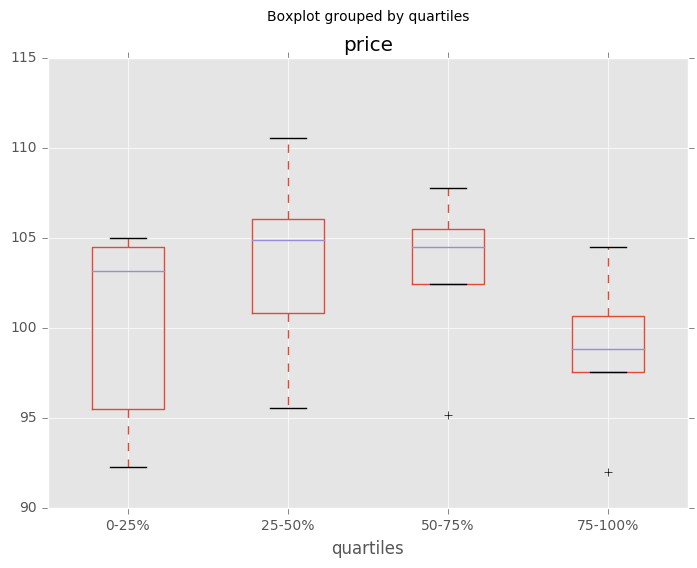
Boxplot for each quartile of a stratifying variable
In [162]: df = pd.DataFrame(
.....: {u'stratifying_var': np.random.uniform(0, 100, 20),
.....: u'price': np.random.normal(100, 5, 20)})
.....:
In [163]: df[u'quartiles'] = pd.qcut(
.....: df[u'stratifying_var'],
.....: 4,
.....: labels=[u'0-25%', u'25-50%', u'50-75%', u'75-100%'])
.....:
In [164]: df.boxplot(column=u'price', by=u'quartiles')
Out[164]: <matplotlib.axes._subplots.AxesSubplot at 0x7fd24d8f8410>
Data In/Out¶
Performance comparison of SQL vs HDF5
CSV¶
The CSV docs
how to read in multiple files, appending to create a single dataframe
Reading only certain rows of a csv chunk-by-chunk
Reading the first few lines of a frame
Reading a file that is compressed but not by gzip/bz2 (the native compressed formats which read_csv understands).
This example shows a WinZipped file, but is a general application of opening the file within a context manager and
using that handle to read.
See here
Reading CSV with Unix timestamps and converting to local timezone
Write a multi-row index CSV without writing duplicates
Parsing date components in multi-columns is faster with a format
In [30]: i = pd.date_range('20000101',periods=10000)
In [31]: df = pd.DataFrame(dict(year = i.year, month = i.month, day = i.day))
In [32]: df.head()
Out[32]:
day month year
0 1 1 2000
1 2 1 2000
2 3 1 2000
3 4 1 2000
4 5 1 2000
In [33]: %timeit pd.to_datetime(df.year*10000+df.month*100+df.day,format='%Y%m%d')
100 loops, best of 3: 7.08 ms per loop
# simulate combinging into a string, then parsing
In [34]: ds = df.apply(lambda x: "%04d%02d%02d" % (x['year'],x['month'],x['day']),axis=1)
In [35]: ds.head()
Out[35]:
0 20000101
1 20000102
2 20000103
3 20000104
4 20000105
dtype: object
In [36]: %timeit pd.to_datetime(ds)
1 loops, best of 3: 488 ms per loop
Skip row between header and data¶
In [165]: from io import StringIO
In [166]: import pandas as pd
In [167]: data = """;;;;
.....: ;;;;
.....: ;;;;
.....: ;;;;
.....: ;;;;
.....: ;;;;
.....: ;;;;
.....: ;;;;
.....: ;;;;
.....: ;;;;
.....: date;Param1;Param2;Param4;Param5
.....: ;m²;°C;m²;m
.....: ;;;;
.....: 01.01.1990 00:00;1;1;2;3
.....: 01.01.1990 01:00;5;3;4;5
.....: 01.01.1990 02:00;9;5;6;7
.....: 01.01.1990 03:00;13;7;8;9
.....: 01.01.1990 04:00;17;9;10;11
.....: 01.01.1990 05:00;21;11;12;13
.....: """
.....:
Option 1: pass rows explicitly to skiprows¶
In [168]: pd.read_csv(StringIO(data.decode('UTF-8')), sep=';', skiprows=[11,12],
.....: index_col=0, parse_dates=True, header=10)
.....:
Out[168]:
Param1 Param2 Param4 Param5
date
1990-01-01 00:00:00 1 1 2 3
1990-01-01 01:00:00 5 3 4 5
1990-01-01 02:00:00 9 5 6 7
1990-01-01 03:00:00 13 7 8 9
1990-01-01 04:00:00 17 9 10 11
1990-01-01 05:00:00 21 11 12 13
Option 2: read column names and then data¶
In [169]: pd.read_csv(StringIO(data.decode('UTF-8')), sep=';',
.....: header=10, parse_dates=True, nrows=10).columns
.....:
Out[169]: Index([u'date', u'Param1', u'Param2', u'Param4', u'Param5'], dtype='object')
In [170]: columns = pd.read_csv(StringIO(data.decode('UTF-8')), sep=';',
.....: header=10, parse_dates=True, nrows=10).columns
.....:
In [171]: pd.read_csv(StringIO(data.decode('UTF-8')), sep=';',
.....: header=12, parse_dates=True, names=columns)
.....:
Out[171]:
date Param1 Param2 Param4 Param5
0 01.01.1990 00:00 1 1 2 3
1 01.01.1990 01:00 5 3 4 5
2 01.01.1990 02:00 9 5 6 7
3 01.01.1990 03:00 13 7 8 9
4 01.01.1990 04:00 17 9 10 11
5 01.01.1990 05:00 21 11 12 13
HDFStore¶
The HDFStores docs
Simple Queries with a Timestamp Index
Managing heterogeneous data using a linked multiple table hierarchy
Merging on-disk tables with millions of rows
Avoiding inconsistencies when writing to a store from multiple processes/threads
De-duplicating a large store by chunks, essentially a recursive reduction operation. Shows a function for taking in data from csv file and creating a store by chunks, with date parsing as well. See here
Creating a store chunk-by-chunk from a csv file
Appending to a store, while creating a unique index
Reading in a sequence of files, then providing a global unique index to a store while appending
Groupby on a HDFStore with low group density
Groupby on a HDFStore with high group density
Hierarchical queries on a HDFStore
Troubleshoot HDFStore exceptions
Setting min_itemsize with strings
Using ptrepack to create a completely-sorted-index on a store
Storing Attributes to a group node
In [172]: df = pd.DataFrame(np.random.randn(8,3))
In [173]: store = pd.HDFStore('test.h5')
In [174]: store.put('df',df)
# you can store an arbitrary python object via pickle
In [175]: store.get_storer('df').attrs.my_attribute = dict(A = 10)
In [176]: store.get_storer('df').attrs.my_attribute
Out[176]: {'A': 10}
Binary Files¶
pandas readily accepts numpy record arrays, if you need to read in a binary
file consisting of an array of C structs. For example, given this C program
in a file called main.c compiled with gcc main.c -std=gnu99 on a
64-bit machine,
#include <stdio.h>
#include <stdint.h>
typedef struct _Data
{
int32_t count;
double avg;
float scale;
} Data;
int main(int argc, const char *argv[])
{
size_t n = 10;
Data d[n];
for (int i = 0; i < n; ++i)
{
d[i].count = i;
d[i].avg = i + 1.0;
d[i].scale = (float) i + 2.0f;
}
FILE *file = fopen("binary.dat", "wb");
fwrite(&d, sizeof(Data), n, file);
fclose(file);
return 0;
}
the following Python code will read the binary file 'binary.dat' into a
pandas DataFrame, where each element of the struct corresponds to a column
in the frame:
names = 'count', 'avg', 'scale'
# note that the offsets are larger than the size of the type because of
# struct padding
offsets = 0, 8, 16
formats = 'i4', 'f8', 'f4'
dt = np.dtype({'names': names, 'offsets': offsets, 'formats': formats},
align=True)
df = pd.DataFrame(np.fromfile('binary.dat', dt))
Note
The offsets of the structure elements may be different depending on the architecture of the machine on which the file was created. Using a raw binary file format like this for general data storage is not recommended, as it is not cross platform. We recommended either HDF5 or msgpack, both of which are supported by pandas’ IO facilities.
Timedeltas¶
The Timedeltas docs.
In [177]: s = pd.Series(pd.date_range('2012-1-1', periods=3, freq='D'))
In [178]: s - s.max()
Out[178]:
0 -2 days
1 -1 days
2 0 days
dtype: timedelta64[ns]
In [179]: s.max() - s
Out[179]:
0 2 days
1 1 days
2 0 days
dtype: timedelta64[ns]
In [180]: s - datetime.datetime(2011,1,1,3,5)
Out[180]:
0 364 days 20:55:00
1 365 days 20:55:00
2 366 days 20:55:00
dtype: timedelta64[ns]
In [181]: s + datetime.timedelta(minutes=5)
Out[181]:
0 2012-01-01 00:05:00
1 2012-01-02 00:05:00
2 2012-01-03 00:05:00
dtype: datetime64[ns]
In [182]: datetime.datetime(2011,1,1,3,5) - s
Out[182]:
0 -365 days +03:05:00
1 -366 days +03:05:00
2 -367 days +03:05:00
dtype: timedelta64[ns]
In [183]: datetime.timedelta(minutes=5) + s
Out[183]:
0 2012-01-01 00:05:00
1 2012-01-02 00:05:00
2 2012-01-03 00:05:00
dtype: datetime64[ns]
Adding and subtracting deltas and dates
In [184]: deltas = pd.Series([ datetime.timedelta(days=i) for i in range(3) ])
In [185]: df = pd.DataFrame(dict(A = s, B = deltas)); df
Out[185]:
A B
0 2012-01-01 0 days
1 2012-01-02 1 days
2 2012-01-03 2 days
In [186]: df['New Dates'] = df['A'] + df['B'];
In [187]: df['Delta'] = df['A'] - df['New Dates']; df
Out[187]:
A B New Dates Delta
0 2012-01-01 0 days 2012-01-01 0 days
1 2012-01-02 1 days 2012-01-03 -1 days
2 2012-01-03 2 days 2012-01-05 -2 days
In [188]: df.dtypes
Out[188]:
A datetime64[ns]
B timedelta64[ns]
New Dates datetime64[ns]
Delta timedelta64[ns]
dtype: object
Values can be set to NaT using np.nan, similar to datetime
In [189]: y = s - s.shift(); y
Out[189]:
0 NaT
1 1 days
2 1 days
dtype: timedelta64[ns]
In [190]: y[1] = np.nan; y
Out[190]:
0 NaT
1 NaT
2 1 days
dtype: timedelta64[ns]
Aliasing Axis Names¶
To globally provide aliases for axis names, one can define these 2 functions:
In [191]: def set_axis_alias(cls, axis, alias):
.....: if axis not in cls._AXIS_NUMBERS:
.....: raise Exception("invalid axis [%s] for alias [%s]" % (axis, alias))
.....: cls._AXIS_ALIASES[alias] = axis
.....:
In [192]: def clear_axis_alias(cls, axis, alias):
.....: if axis not in cls._AXIS_NUMBERS:
.....: raise Exception("invalid axis [%s] for alias [%s]" % (axis, alias))
.....: cls._AXIS_ALIASES.pop(alias,None)
.....:
In [193]: set_axis_alias(pd.DataFrame,'columns', 'myaxis2')
In [194]: df2 = pd.DataFrame(np.random.randn(3,2),columns=['c1','c2'],index=['i1','i2','i3'])
In [195]: df2.sum(axis='myaxis2')
Out[195]:
i1 -0.573143
i2 -0.161663
i3 0.264035
dtype: float64
In [196]: clear_axis_alias(pd.DataFrame,'columns', 'myaxis2')
Creating Example Data¶
To create a dataframe from every combination of some given values, like R’s expand.grid()
function, we can create a dict where the keys are column names and the values are lists
of the data values:
In [197]: def expand_grid(data_dict):
.....: rows = itertools.product(*data_dict.values())
.....: return pd.DataFrame.from_records(rows, columns=data_dict.keys())
.....:
In [198]: df = expand_grid(
.....: {'height': [60, 70],
.....: 'weight': [100, 140, 180],
.....: 'sex': ['Male', 'Female']})
.....:
In [199]: df
Out[199]:
sex weight height
0 Male 100 60
1 Male 100 70
2 Male 140 60
3 Male 140 70
4 Male 180 60
5 Male 180 70
6 Female 100 60
7 Female 100 70
8 Female 140 60
9 Female 140 70
10 Female 180 60
11 Female 180 70