Contributing to the code base¶
Table of Contents:
Code standards¶
Writing good code is not just about what you write. It is also about how you write it. During Continuous Integration testing, several tools will be run to check your code for stylistic errors. Generating any warnings will cause the test to fail. Thus, good style is a requirement for submitting code to pandas.
There is a tool in pandas to help contributors verify their changes before contributing them to the project:
./ci/code_checks.sh
The script verifies the linting of code files, it looks for common mistake patterns
(like missing spaces around sphinx directives that make the documentation not
being rendered properly) and it also validates the doctests. It is possible to
run the checks independently by using the parameters lint, patterns and
doctests (e.g. ./ci/code_checks.sh lint).
In addition, because a lot of people use our library, it is important that we do not make sudden changes to the code that could have the potential to break a lot of user code as a result, that is, we need it to be as backwards compatible as possible to avoid mass breakages.
In addition to ./ci/code_checks.sh, some extra checks are run by
pre-commit - see here for how to
run them.
Additional standards are outlined on the pandas code style guide.
Pre-commit¶
You can run many of these styling checks manually as we have described above. However,
we encourage you to use pre-commit hooks instead
to automatically run black, flake8, isort when you make a git commit. This
can be done by installing pre-commit:
pip install pre-commit
and then running:
pre-commit install
from the root of the pandas repository. Now all of the styling checks will be
run each time you commit changes without your needing to run each one manually.
In addition, using pre-commit will also allow you to more easily
remain up-to-date with our code checks as they change.
Note that if needed, you can skip these checks with git commit --no-verify.
If you don’t want to use pre-commit as part of your workflow, you can still use it
to run its checks with:
pre-commit run --files <files you have modified>
without needing to have done pre-commit install beforehand.
If you want to run checks on all recently committed files on upstream/master you can use:
pre-commit run --from-ref=upstream/master --to-ref=HEAD --all-files
without needing to have done pre-commit install beforehand.
Note
If you have conflicting installations of virtualenv, then you may get an
error - see here.
Also, due to a bug in virtualenv,
you may run into issues if you’re using conda. To solve this, you can downgrade
virtualenv to version 20.0.33.
Optional dependencies¶
Optional dependencies (e.g. matplotlib) should be imported with the private helper
pandas.compat._optional.import_optional_dependency. This ensures a
consistent error message when the dependency is not met.
All methods using an optional dependency should include a test asserting that an
ImportError is raised when the optional dependency is not found. This test
should be skipped if the library is present.
All optional dependencies should be documented in
Optional dependencies and the minimum required version should be
set in the pandas.compat._optional.VERSIONS dict.
C (cpplint)¶
pandas uses the Google
standard. Google provides an open source style checker called cpplint, but we
use a fork of it that can be found here.
Here are some of the more common cpplint issues:
we restrict line-length to 80 characters to promote readability
every header file must include a header guard to avoid name collisions if re-included
Continuous Integration will run the cpplint tool and report any stylistic errors in your code. Therefore, it is helpful before submitting code to run the check yourself:
cpplint --extensions=c,h --headers=h --filter=-readability/casting,-runtime/int,-build/include_subdir modified-c-file
You can also run this command on an entire directory if necessary:
cpplint --extensions=c,h --headers=h --filter=-readability/casting,-runtime/int,-build/include_subdir --recursive modified-c-directory
To make your commits compliant with this standard, you can install the ClangFormat tool, which can be downloaded here. To configure, in your home directory, run the following command:
clang-format style=google -dump-config > .clang-format
Then modify the file to ensure that any indentation width parameters are at least four. Once configured, you can run the tool as follows:
clang-format modified-c-file
This will output what your file will look like if the changes are made, and to apply them, run the following command:
clang-format -i modified-c-file
To run the tool on an entire directory, you can run the following analogous commands:
clang-format modified-c-directory/*.c modified-c-directory/*.h
clang-format -i modified-c-directory/*.c modified-c-directory/*.h
Do note that this tool is best-effort, meaning that it will try to correct as
many errors as possible, but it may not correct all of them. Thus, it is
recommended that you run cpplint to double check and make any other style
fixes manually.
Python (PEP8 / black)¶
pandas follows the PEP8 standard and uses Black and Flake8 to ensure a consistent code format throughout the project. We encourage you to use pre-commit.
Continuous Integration will run those tools and report any stylistic errors in your code. Therefore, it is helpful before submitting code to run the check yourself:
black pandas
git diff upstream/master -u -- "*.py" | flake8 --diff
to auto-format your code. Additionally, many editors have plugins that will
apply black as you edit files.
You should use a black version 21.5b2 as previous versions are not compatible
with the pandas codebase.
One caveat about git diff upstream/master -u -- "*.py" | flake8 --diff: this
command will catch any stylistic errors in your changes specifically, but
be beware it may not catch all of them. For example, if you delete the only
usage of an imported function, it is stylistically incorrect to import an
unused function. However, style-checking the diff will not catch this because
the actual import is not part of the diff. Thus, for completeness, you should
run this command, though it may take longer:
git diff upstream/master --name-only -- "*.py" | xargs -r flake8
Note that on OSX, the -r flag is not available, so you have to omit it and
run this slightly modified command:
git diff upstream/master --name-only -- "*.py" | xargs flake8
Windows does not support the xargs command (unless installed for example
via the MinGW toolchain), but one can imitate the
behaviour as follows:
for /f %i in ('git diff upstream/master --name-only -- "*.py"') do flake8 %i
This will get all the files being changed by the PR (and ending with .py),
and run flake8 on them, one after the other.
Note that these commands can be run analogously with black.
Import formatting¶
pandas uses isort to standardise import formatting across the codebase.
A guide to import layout as per pep8 can be found here.
A summary of our current import sections ( in order ):
Future
Python Standard Library
Third Party
pandas._libs,pandas.compat,pandas.util._*,pandas.errors(largely not dependent onpandas.core)pandas.core.dtypes(largely not dependent on the rest ofpandas.core)Rest of
pandas.core.*Non-core
pandas.io,pandas.plotting,pandas.tseriesLocal application/library specific imports
Imports are alphabetically sorted within these sections.
As part of Continuous Integration checks we run:
isort --check-only pandas
to check that imports are correctly formatted as per the setup.cfg.
If you see output like the below in Continuous Integration checks:
Check import format using isort
ERROR: /home/travis/build/pandas-dev/pandas/pandas/io/pytables.py Imports are incorrectly sorted
Check import format using isort DONE
The command "ci/code_checks.sh" exited with 1
You should run:
isort pandas/io/pytables.py
to automatically format imports correctly. This will modify your local copy of the files.
Alternatively, you can run a command similar to what was suggested for black and flake8 right above:
git diff upstream/master --name-only -- "*.py" | xargs -r isort
Where similar caveats apply if you are on OSX or Windows.
You can then verify the changes look ok, then git commit and push.
Backwards compatibility¶
Please try to maintain backward compatibility. pandas has lots of users with lots of existing code, so don’t break it if at all possible. If you think breakage is required, clearly state why as part of the pull request. Also, be careful when changing method signatures and add deprecation warnings where needed. Also, add the deprecated sphinx directive to the deprecated functions or methods.
If a function with the same arguments as the one being deprecated exist, you can use
the pandas.util._decorators.deprecate:
from pandas.util._decorators import deprecate
deprecate('old_func', 'new_func', '1.1.0')
Otherwise, you need to do it manually:
import warnings
def old_func():
"""Summary of the function.
.. deprecated:: 1.1.0
Use new_func instead.
"""
warnings.warn('Use new_func instead.', FutureWarning, stacklevel=2)
new_func()
def new_func():
pass
You’ll also need to
Write a new test that asserts a warning is issued when calling with the deprecated argument
Update all of pandas existing tests and code to use the new argument
See Testing warnings for more.
Type hints¶
pandas strongly encourages the use of PEP 484 style type hints. New development should contain type hints and pull requests to annotate existing code are accepted as well!
Style guidelines¶
Types imports should follow the from typing import ... convention. So rather than
import typing
primes: typing.List[int] = []
You should write
from typing import List, Optional, Union
primes: List[int] = []
Optional should be used where applicable, so instead of
maybe_primes: List[Union[int, None]] = []
You should write
maybe_primes: List[Optional[int]] = []
In some cases in the code base classes may define class variables that shadow builtins. This causes an issue as described in Mypy 1775. The defensive solution here is to create an unambiguous alias of the builtin and use that without your annotation. For example, if you come across a definition like
class SomeClass1:
str = None
The appropriate way to annotate this would be as follows
str_type = str
class SomeClass2:
str: str_type = None
In some cases you may be tempted to use cast from the typing module when you know better than the analyzer. This occurs particularly when using custom inference functions. For example
from typing import cast
from pandas.core.dtypes.common import is_number
def cannot_infer_bad(obj: Union[str, int, float]):
if is_number(obj):
...
else: # Reasonably only str objects would reach this but...
obj = cast(str, obj) # Mypy complains without this!
return obj.upper()
The limitation here is that while a human can reasonably understand that is_number would catch the int and float types mypy cannot make that same inference just yet (see mypy #5206. While the above works, the use of cast is strongly discouraged. Where applicable a refactor of the code to appease static analysis is preferable
def cannot_infer_good(obj: Union[str, int, float]):
if isinstance(obj, str):
return obj.upper()
else:
...
With custom types and inference this is not always possible so exceptions are made, but every effort should be exhausted to avoid cast before going down such paths.
pandas-specific types¶
Commonly used types specific to pandas will appear in pandas._typing and you should use these where applicable. This module is private for now but ultimately this should be exposed to third party libraries who want to implement type checking against pandas.
For example, quite a few functions in pandas accept a dtype argument. This can be expressed as a string like "object", a numpy.dtype like np.int64 or even a pandas ExtensionDtype like pd.CategoricalDtype. Rather than burden the user with having to constantly annotate all of those options, this can simply be imported and reused from the pandas._typing module
from pandas._typing import Dtype
def as_type(dtype: Dtype) -> ...:
...
This module will ultimately house types for repeatedly used concepts like “path-like”, “array-like”, “numeric”, etc… and can also hold aliases for commonly appearing parameters like axis. Development of this module is active so be sure to refer to the source for the most up to date list of available types.
Validating type hints¶
pandas uses mypy to statically analyze the code base and type hints. After making any change you can ensure your type hints are correct by running
mypy pandas
Testing with continuous integration¶
The pandas test suite will run automatically on GitHub Actions and Azure Pipelines continuous integration services, once your pull request is submitted. However, if you wish to run the test suite on a branch prior to submitting the pull request, then the continuous integration services need to be hooked to your GitHub repository. Instructions are here for GitHub Actions and Azure Pipelines.
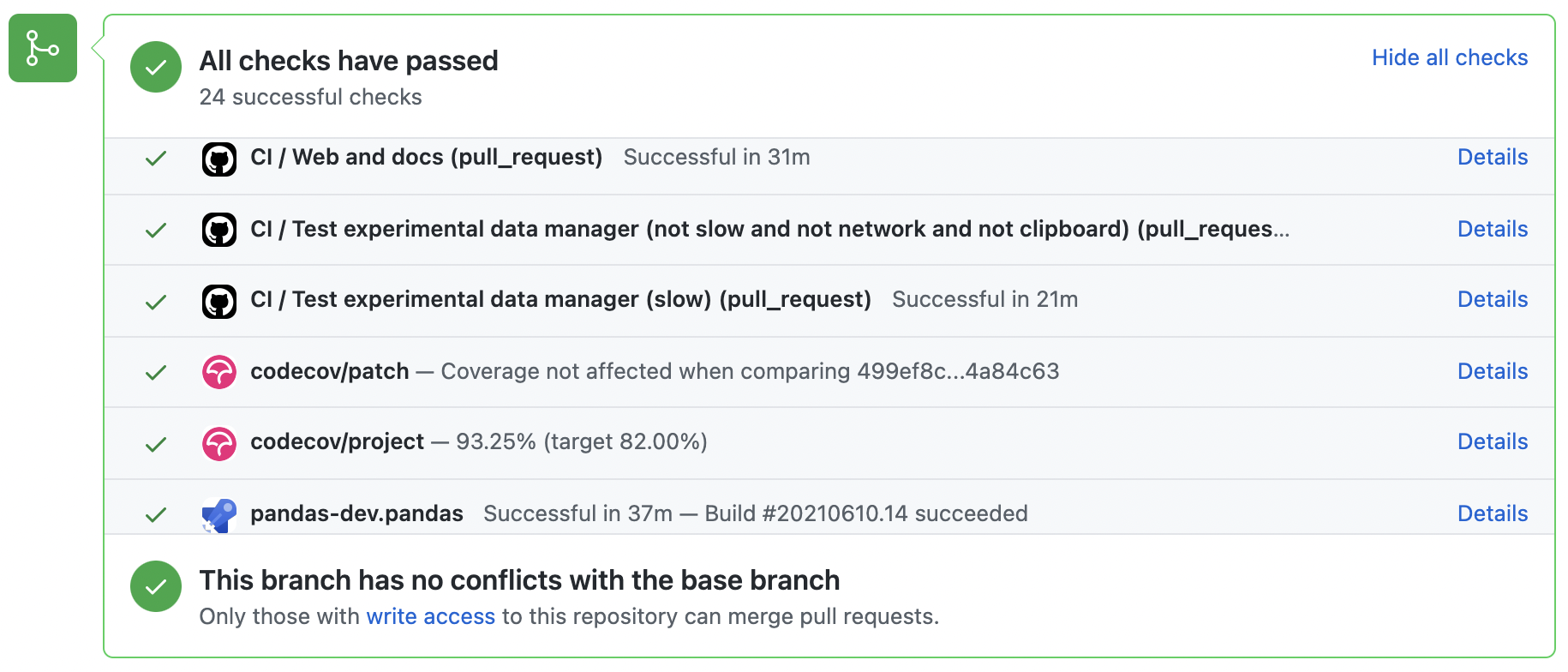
A pull-request will be considered for merging when you have an all ‘green’ build. If any tests are failing, then you will get a red ‘X’, where you can click through to see the individual failed tests. This is an example of a green build.
Test-driven development/code writing¶
pandas is serious about testing and strongly encourages contributors to embrace test-driven development (TDD). This development process “relies on the repetition of a very short development cycle: first the developer writes an (initially failing) automated test case that defines a desired improvement or new function, then produces the minimum amount of code to pass that test.” So, before actually writing any code, you should write your tests. Often the test can be taken from the original GitHub issue. However, it is always worth considering additional use cases and writing corresponding tests.
Adding tests is one of the most common requests after code is pushed to pandas. Therefore, it is worth getting in the habit of writing tests ahead of time so this is never an issue.
Like many packages, pandas uses pytest and the convenient extensions in numpy.testing.
Note
The earliest supported pytest version is 5.0.1.
Writing tests¶
All tests should go into the tests subdirectory of the specific package.
This folder contains many current examples of tests, and we suggest looking to these for
inspiration. If your test requires working with files or
network connectivity, there is more information on the testing page of the wiki.
The pandas._testing module has many special assert functions that
make it easier to make statements about whether Series or DataFrame objects are
equivalent. The easiest way to verify that your code is correct is to
explicitly construct the result you expect, then compare the actual result to
the expected correct result:
def test_pivot(self):
data = {
'index' : ['A', 'B', 'C', 'C', 'B', 'A'],
'columns' : ['One', 'One', 'One', 'Two', 'Two', 'Two'],
'values' : [1., 2., 3., 3., 2., 1.]
}
frame = DataFrame(data)
pivoted = frame.pivot(index='index', columns='columns', values='values')
expected = DataFrame({
'One' : {'A' : 1., 'B' : 2., 'C' : 3.},
'Two' : {'A' : 1., 'B' : 2., 'C' : 3.}
})
assert_frame_equal(pivoted, expected)
Please remember to add the Github Issue Number as a comment to a new test. E.g. “# brief comment, see GH#28907”
Transitioning to pytest¶
pandas existing test structure is mostly class-based, meaning that you will typically find tests wrapped in a class.
class TestReallyCoolFeature:
pass
Going forward, we are moving to a more functional style using the pytest framework, which offers a richer testing framework that will facilitate testing and developing. Thus, instead of writing test classes, we will write test functions like this:
def test_really_cool_feature():
pass
Using pytest¶
Here is an example of a self-contained set of tests that illustrate multiple features that we like to use.
functional style: tests are like
test_*and only take arguments that are either fixtures or parameterspytest.markcan be used to set metadata on test functions, e.g.skiporxfail.using
parametrize: allow testing of multiple casesto set a mark on a parameter,
pytest.param(..., marks=...)syntax should be usedfixture, code for object construction, on a per-test basisusing bare
assertfor scalars and truth-testingtm.assert_series_equal(and its counter parttm.assert_frame_equal), for pandas object comparisons.the typical pattern of constructing an
expectedand comparing versus theresult
We would name this file test_cool_feature.py and put in an appropriate place in the pandas/tests/ structure.
import pytest
import numpy as np
import pandas as pd
@pytest.mark.parametrize('dtype', ['int8', 'int16', 'int32', 'int64'])
def test_dtypes(dtype):
assert str(np.dtype(dtype)) == dtype
@pytest.mark.parametrize(
'dtype', ['float32', pytest.param('int16', marks=pytest.mark.skip),
pytest.param('int32', marks=pytest.mark.xfail(
reason='to show how it works'))])
def test_mark(dtype):
assert str(np.dtype(dtype)) == 'float32'
@pytest.fixture
def series():
return pd.Series([1, 2, 3])
@pytest.fixture(params=['int8', 'int16', 'int32', 'int64'])
def dtype(request):
return request.param
def test_series(series, dtype):
result = series.astype(dtype)
assert result.dtype == dtype
expected = pd.Series([1, 2, 3], dtype=dtype)
tm.assert_series_equal(result, expected)
A test run of this yields
((pandas) bash-3.2$ pytest test_cool_feature.py -v
=========================== test session starts ===========================
platform darwin -- Python 3.6.2, pytest-3.6.0, py-1.4.31, pluggy-0.4.0
collected 11 items
tester.py::test_dtypes[int8] PASSED
tester.py::test_dtypes[int16] PASSED
tester.py::test_dtypes[int32] PASSED
tester.py::test_dtypes[int64] PASSED
tester.py::test_mark[float32] PASSED
tester.py::test_mark[int16] SKIPPED
tester.py::test_mark[int32] xfail
tester.py::test_series[int8] PASSED
tester.py::test_series[int16] PASSED
tester.py::test_series[int32] PASSED
tester.py::test_series[int64] PASSED
Tests that we have parametrized are now accessible via the test name, for example we could run these with -k int8 to sub-select only those tests which match int8.
((pandas) bash-3.2$ pytest test_cool_feature.py -v -k int8
=========================== test session starts ===========================
platform darwin -- Python 3.6.2, pytest-3.6.0, py-1.4.31, pluggy-0.4.0
collected 11 items
test_cool_feature.py::test_dtypes[int8] PASSED
test_cool_feature.py::test_series[int8] PASSED
Using hypothesis¶
Hypothesis is a library for property-based testing. Instead of explicitly parametrizing a test, you can describe all valid inputs and let Hypothesis try to find a failing input. Even better, no matter how many random examples it tries, Hypothesis always reports a single minimal counterexample to your assertions - often an example that you would never have thought to test.
See Getting Started with Hypothesis for more of an introduction, then refer to the Hypothesis documentation for details.
import json
from hypothesis import given, strategies as st
any_json_value = st.deferred(lambda: st.one_of(
st.none(), st.booleans(), st.floats(allow_nan=False), st.text(),
st.lists(any_json_value), st.dictionaries(st.text(), any_json_value)
))
@given(value=any_json_value)
def test_json_roundtrip(value):
result = json.loads(json.dumps(value))
assert value == result
This test shows off several useful features of Hypothesis, as well as demonstrating a good use-case: checking properties that should hold over a large or complicated domain of inputs.
To keep the pandas test suite running quickly, parametrized tests are preferred if the inputs or logic are simple, with Hypothesis tests reserved for cases with complex logic or where there are too many combinations of options or subtle interactions to test (or think of!) all of them.
Testing warnings¶
By default, one of pandas CI workers will fail if any unhandled warnings are emitted.
If your change involves checking that a warning is actually emitted, use
tm.assert_produces_warning(ExpectedWarning).
import pandas._testing as tm
df = pd.DataFrame()
with tm.assert_produces_warning(FutureWarning):
df.some_operation()
We prefer this to the pytest.warns context manager because ours checks that the warning’s
stacklevel is set correctly. The stacklevel is what ensure the user’s file name and line number
is printed in the warning, rather than something internal to pandas. It represents the number of
function calls from user code (e.g. df.some_operation()) to the function that actually emits
the warning. Our linter will fail the build if you use pytest.warns in a test.
If you have a test that would emit a warning, but you aren’t actually testing the
warning itself (say because it’s going to be removed in the future, or because we’re
matching a 3rd-party library’s behavior), then use pytest.mark.filterwarnings to
ignore the error.
@pytest.mark.filterwarnings("ignore:msg:category")
def test_thing(self):
...
If the test generates a warning of class category whose message starts
with msg, the warning will be ignored and the test will pass.
If you need finer-grained control, you can use Python’s usual warnings module to control whether a warning is ignored / raised at different places within a single test.
with warnings.catch_warnings():
warnings.simplefilter("ignore", FutureWarning)
# Or use warnings.filterwarnings(...)
Alternatively, consider breaking up the unit test.
Running the test suite¶
The tests can then be run directly inside your Git clone (without having to install pandas) by typing:
pytest pandas
The tests suite is exhaustive and takes around 20 minutes to run. Often it is worth running only a subset of tests first around your changes before running the entire suite.
The easiest way to do this is with:
pytest pandas/path/to/test.py -k regex_matching_test_name
Or with one of the following constructs:
pytest pandas/tests/[test-module].py
pytest pandas/tests/[test-module].py::[TestClass]
pytest pandas/tests/[test-module].py::[TestClass]::[test_method]
Using pytest-xdist, one can
speed up local testing on multicore machines. To use this feature, you will
need to install pytest-xdist via:
pip install pytest-xdist
Two scripts are provided to assist with this. These scripts distribute testing across 4 threads.
On Unix variants, one can type:
test_fast.sh
On Windows, one can type:
test_fast.bat
This can significantly reduce the time it takes to locally run tests before submitting a pull request.
For more, see the pytest documentation.
Furthermore one can run
pd.test()
with an imported pandas to run tests similarly.
Running the performance test suite¶
Performance matters and it is worth considering whether your code has introduced
performance regressions. pandas is in the process of migrating to
asv benchmarks
to enable easy monitoring of the performance of critical pandas operations.
These benchmarks are all found in the pandas/asv_bench directory, and the
test results can be found here.
To use all features of asv, you will need either conda or
virtualenv. For more details please check the asv installation
webpage.
To install asv:
pip install git+https://github.com/spacetelescope/asv
If you need to run a benchmark, change your directory to asv_bench/ and run:
asv continuous -f 1.1 upstream/master HEAD
You can replace HEAD with the name of the branch you are working on,
and report benchmarks that changed by more than 10%.
The command uses conda by default for creating the benchmark
environments. If you want to use virtualenv instead, write:
asv continuous -f 1.1 -E virtualenv upstream/master HEAD
The -E virtualenv option should be added to all asv commands
that run benchmarks. The default value is defined in asv.conf.json.
Running the full benchmark suite can be an all-day process, depending on your
hardware and its resource utilization. However, usually it is sufficient to paste
only a subset of the results into the pull request to show that the committed changes
do not cause unexpected performance regressions. You can run specific benchmarks
using the -b flag, which takes a regular expression. For example, this will
only run benchmarks from a pandas/asv_bench/benchmarks/groupby.py file:
asv continuous -f 1.1 upstream/master HEAD -b ^groupby
If you want to only run a specific group of benchmarks from a file, you can do it
using . as a separator. For example:
asv continuous -f 1.1 upstream/master HEAD -b groupby.GroupByMethods
will only run the GroupByMethods benchmark defined in groupby.py.
You can also run the benchmark suite using the version of pandas
already installed in your current Python environment. This can be
useful if you do not have virtualenv or conda, or are using the
setup.py develop approach discussed above; for the in-place build
you need to set PYTHONPATH, e.g.
PYTHONPATH="$PWD/.." asv [remaining arguments].
You can run benchmarks using an existing Python
environment by:
asv run -e -E existing
or, to use a specific Python interpreter,:
asv run -e -E existing:python3.6
This will display stderr from the benchmarks, and use your local
python that comes from your $PATH.
Information on how to write a benchmark and how to use asv can be found in the asv documentation.
Documenting your code¶
Changes should be reflected in the release notes located in doc/source/whatsnew/vx.y.z.rst.
This file contains an ongoing change log for each release. Add an entry to this file to
document your fix, enhancement or (unavoidable) breaking change. Make sure to include the
GitHub issue number when adding your entry (using :issue:`1234` where 1234 is the
issue/pull request number).
If your code is an enhancement, it is most likely necessary to add usage
examples to the existing documentation. This can be done following the section
regarding documentation.
Further, to let users know when this feature was added, the versionadded
directive is used. The sphinx syntax for that is:
.. versionadded:: 1.1.0
This will put the text New in version 1.1.0 wherever you put the sphinx directive. This should also be put in the docstring when adding a new function or method (example) or a new keyword argument (example).