What’s New¶
These are new features and improvements of note in each release.
v0.20.3 (July 7, 2017)¶
This is a minor bug-fix release in the 0.20.x series and includes some small regression fixes and bug fixes. We recommend that all users upgrade to this version.
What’s new in v0.20.3
Bug Fixes¶
- Fixed a bug in failing to compute rolling computations of a column-MultiIndexed
DataFrame(GH16789, GH16825) - Fixed a pytest marker failing downstream packages’ tests suites (GH16680)
Conversion¶
- Bug in pickle compat prior to the v0.20.x series, when
UTCis a timezone in a Series/DataFrame/Index (GH16608) - Bug in
Seriesconstruction when passing aSerieswithdtype='category'(GH16524). - Bug in
DataFrame.astype()when passing aSeriesas thedtypekwarg. (GH16717).
Indexing¶
- Bug in
Float64Indexcausing an empty array instead ofNoneto be returned from.get(np.nan)on a Series whose index did not contain anyNaNs (GH8569) - Bug in
MultiIndex.isincausing an error when passing an empty iterable (GH16777) - Fixed a bug in a slicing DataFrame/Series that have a
TimedeltaIndex(GH16637)
I/O¶
- Bug in
read_csv()in which files weren’t opened as binary files by the C engine on Windows, causing EOF characters mid-field, which would fail (GH16039, GH16559, GH16675) - Bug in
read_hdf()in which reading aSeriessaved to an HDF file in ‘fixed’ format fails when an explicitmode='r'argument is supplied (GH16583) - Bug in
DataFrame.to_latex()wherebold_rowswas wrongly specified to beTrueby default, whereas in reality row labels remained non-bold whatever parameter provided. (GH16707) - Fixed an issue with
DataFrame.style()where generated element ids were not unique (GH16780) - Fixed loading a
DataFramewith aPeriodIndex, from aformat='fixed'HDFStore, in Python 3, that was written in Python 2 (GH16781)
Plotting¶
- Fixed regression that prevented RGB and RGBA tuples from being used as color arguments (GH16233)
- Fixed an issue with
DataFrame.plot.scatter()that incorrectly raised aKeyErrorwhen categorical data is used for plotting (GH16199)
Reshaping¶
v0.20.2 (June 4, 2017)¶
This is a minor bug-fix release in the 0.20.x series and includes some small regression fixes, bug fixes and performance improvements. We recommend that all users upgrade to this version.
What’s new in v0.20.2
Enhancements¶
- Unblocked access to additional compression types supported in pytables: ‘blosc:blosclz, ‘blosc:lz4’, ‘blosc:lz4hc’, ‘blosc:snappy’, ‘blosc:zlib’, ‘blosc:zstd’ (GH14478)
Seriesprovides ato_latexmethod (GH16180)- A new groupby method
ngroup(), parallel to the existingcumcount(), has been added to return the group order (GH11642); see here.
Performance Improvements¶
- Performance regression fix when indexing with a list-like (GH16285)
- Performance regression fix for MultiIndexes (GH16319, GH16346)
- Improved performance of
.clip()with scalar arguments (GH15400) - Improved performance of groupby with categorical groupers (GH16413)
- Improved performance of
MultiIndex.remove_unused_levels()(GH16556)
Bug Fixes¶
- Silenced a warning on some Windows environments about “tput: terminal attributes: No such device or address” when detecting the terminal size. This fix only applies to python 3 (GH16496)
- Bug in using
pathlib.Pathorpy.path.localobjects with io functions (GH16291) - Bug in
Index.symmetric_difference()on two equal MultiIndex’s, results in aTypeError(:issue 13490) - Bug in
DataFrame.update()withoverwrite=FalseandNaN values(GH15593) - Passing an invalid engine to
read_csv()now raises an informativeValueErrorrather thanUnboundLocalError. (GH16511) - Bug in
unique()on an array of tuples (GH16519) - Bug in
cut()whenlabelsare set, resulting in incorrect label ordering (GH16459) - Fixed a compatibility issue with IPython 6.0’s tab completion showing deprecation warnings on
Categoricals(GH16409)
Conversion¶
- Bug in
to_numeric()in which empty data inputs were causing a segfault of the interpreter (GH16302) - Silence numpy warnings when broadcasting
DataFrametoSerieswith comparison ops (GH16378, GH16306)
Indexing¶
- Bug in
DataFrame.reset_index(level=)with single level index (GH16263) - Bug in partial string indexing with a monotonic, but not strictly-monotonic, index incorrectly reversing the slice bounds (GH16515)
- Bug in
MultiIndex.remove_unused_levels()that would not return aMultiIndexequal to the original. (GH16556)
I/O¶
- Bug in
read_csv()whencommentis passed in a space delimited text file (GH16472) - Bug in
read_csv()not raising an exception with nonexistent columns inusecolswhen it had the correct length (GH14671) - Bug that would force importing of the clipboard routines unnecessarily, potentially causing an import error on startup (GH16288)
- Bug that raised
IndexErrorwhen HTML-rendering an emptyDataFrame(GH15953) - Bug in
read_csv()in which tarfile object inputs were raising an error in Python 2.x for the C engine (GH16530) - Bug where
DataFrame.to_html()ignored theindex_namesparameter (GH16493) - Bug where
pd.read_hdf()returns numpy strings for index names (GH13492) - Bug in
HDFStore.select_as_multiple()where start/stop arguments were not respected (GH16209)
Plotting¶
Groupby/Resample/Rolling¶
Reshaping¶
- Bug in
DataFrame.stackwith unsorted levels inMultiIndexcolumns (GH16323) - Bug in
pd.wide_to_long()where no error was raised wheniwas not a unique identifier (GH16382) - Bug in
Series.isin(..)with a list of tuples (GH16394) - Bug in construction of a
DataFramewith mixed dtypes including an all-NaT column. (GH16395) - Bug in
DataFrame.agg()andSeries.agg()with aggregating on non-callable attributes (GH16405)
Numeric¶
- Bug in
.interpolate(), wherelimit_directionwas not respected whenlimit=None(default) was passed (GH16282)
v0.20.1 (May 5, 2017)¶
This is a major release from 0.19.2 and includes a number of API changes, deprecations, new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
Highlights include:
- New
.agg()API for Series/DataFrame similar to the groupby-rolling-resample API’s, see here - Integration with the
feather-format, including a new top-levelpd.read_feather()andDataFrame.to_feather()method, see here. - The
.ixindexer has been deprecated, see here Panelhas been deprecated, see here- Addition of an
IntervalIndexandIntervalscalar type, see here - Improved user API when grouping by index levels in
.groupby(), see here - Improved support for
UInt64dtypes, see here - A new orient for JSON serialization,
orient='table', that uses the Table Schema spec and that gives the possibility for a more interactive repr in the Jupyter Notebook, see here - Experimental support for exporting styled DataFrames (
DataFrame.style) to Excel, see here - Window binary corr/cov operations now return a MultiIndexed
DataFramerather than aPanel, asPanelis now deprecated, see here - Support for S3 handling now uses
s3fs, see here - Google BigQuery support now uses the
pandas-gbqlibrary, see here
Warning
Pandas has changed the internal structure and layout of the codebase.
This can affect imports that are not from the top-level pandas.* namespace, please see the changes here.
Check the API Changes and deprecations before updating.
Note
This is a combined release for 0.20.0 and and 0.20.1.
Version 0.20.1 contains one additional change for backwards-compatibility with downstream projects using pandas’ utils routines. (GH16250)
What’s new in v0.20.0
- New features
aggAPI for DataFrame/Seriesdtypekeyword for data IO.to_datetime()has gained anoriginparameter- Groupby Enhancements
- Better support for compressed URLs in
read_csv - Pickle file I/O now supports compression
- UInt64 Support Improved
- GroupBy on Categoricals
- Table Schema Output
- SciPy sparse matrix from/to SparseDataFrame
- Excel output for styled DataFrames
- IntervalIndex
- Other Enhancements
- Backwards incompatible API changes
- Possible incompatibility for HDF5 formats created with pandas < 0.13.0
- Map on Index types now return other Index types
- Accessing datetime fields of Index now return Index
- pd.unique will now be consistent with extension types
- S3 File Handling
- Partial String Indexing Changes
- Concat of different float dtypes will not automatically upcast
- Pandas Google BigQuery support has moved
- Memory Usage for Index is more Accurate
- DataFrame.sort_index changes
- Groupby Describe Formatting
- Window Binary Corr/Cov operations return a MultiIndex DataFrame
- HDFStore where string comparison
- Index.intersection and inner join now preserve the order of the left Index
- Pivot Table always returns a DataFrame
- Other API Changes
- Reorganization of the library: Privacy Changes
- Deprecations
- Removal of prior version deprecations/changes
- Performance Improvements
- Bug Fixes
New features¶
agg API for DataFrame/Series¶
Series & DataFrame have been enhanced to support the aggregation API. This is a familiar API
from groupby, window operations, and resampling. This allows aggregation operations in a concise way
by using agg() and transform(). The full documentation
is here (GH1623).
Here is a sample
In [1]: df = pd.DataFrame(np.random.randn(10, 3), columns=['A', 'B', 'C'],
...: index=pd.date_range('1/1/2000', periods=10))
...:
In [2]: df.iloc[3:7] = np.nan
In [3]: df
Out[3]:
A B C
2000-01-01 1.474071 -0.064034 -1.282782
2000-01-02 0.781836 -1.071357 0.441153
2000-01-03 2.353925 0.583787 0.221471
2000-01-04 NaN NaN NaN
2000-01-05 NaN NaN NaN
2000-01-06 NaN NaN NaN
2000-01-07 NaN NaN NaN
2000-01-08 0.901805 1.171216 0.520260
2000-01-09 -1.197071 -1.066969 -0.303421
2000-01-10 -0.858447 0.306996 -0.028665
One can operate using string function names, callables, lists, or dictionaries of these.
Using a single function is equivalent to .apply.
In [4]: df.agg('sum')
Out[4]:
A 3.456119
B -0.140361
C -0.431984
dtype: float64
Multiple aggregations with a list of functions.
In [5]: df.agg(['sum', 'min'])
Out[5]:
A B C
sum 3.456119 -0.140361 -0.431984
min -1.197071 -1.071357 -1.282782
Using a dict provides the ability to apply specific aggregations per column.
You will get a matrix-like output of all of the aggregators. The output has one column
per unique function. Those functions applied to a particular column will be NaN:
In [6]: df.agg({'A' : ['sum', 'min'], 'B' : ['min', 'max']})
Out[6]:
A B
max NaN 1.171216
min -1.197071 -1.071357
sum 3.456119 NaN
The API also supports a .transform() function for broadcasting results.
In [7]: df.transform(['abs', lambda x: x - x.min()])
Out[7]:
A B C
abs <lambda> abs <lambda> abs <lambda>
2000-01-01 1.474071 2.671143 0.064034 1.007322 1.282782 0.000000
2000-01-02 0.781836 1.978907 1.071357 0.000000 0.441153 1.723935
2000-01-03 2.353925 3.550996 0.583787 1.655143 0.221471 1.504252
2000-01-04 NaN NaN NaN NaN NaN NaN
2000-01-05 NaN NaN NaN NaN NaN NaN
2000-01-06 NaN NaN NaN NaN NaN NaN
2000-01-07 NaN NaN NaN NaN NaN NaN
2000-01-08 0.901805 2.098877 1.171216 2.242573 0.520260 1.803042
2000-01-09 1.197071 0.000000 1.066969 0.004388 0.303421 0.979361
2000-01-10 0.858447 0.338624 0.306996 1.378353 0.028665 1.254117
When presented with mixed dtypes that cannot be aggregated, .agg() will only take the valid
aggregations. This is similiar to how groupby .agg() works. (GH15015)
In [8]: df = pd.DataFrame({'A': [1, 2, 3],
...: 'B': [1., 2., 3.],
...: 'C': ['foo', 'bar', 'baz'],
...: 'D': pd.date_range('20130101', periods=3)})
...:
In [9]: df.dtypes
Out[9]:
A int64
B float64
C object
D datetime64[ns]
dtype: object
In [10]: df.agg(['min', 'sum'])
Out[10]:
A B C D
min 1 1.0 bar 2013-01-01
sum 6 6.0 foobarbaz NaT
dtype keyword for data IO¶
The 'python' engine for read_csv(), as well as the read_fwf() function for parsing
fixed-width text files and read_excel() for parsing Excel files, now accept the dtype keyword argument for specifying the types of specific columns (GH14295). See the io docs for more information.
In [11]: data = "a b\n1 2\n3 4"
In [12]: pd.read_fwf(StringIO(data)).dtypes
Out[12]:
a int64
b int64
dtype: object
In [13]: pd.read_fwf(StringIO(data), dtype={'a':'float64', 'b':'object'}).dtypes
����������������������������������������������Out[13]:
a float64
b object
dtype: object
.to_datetime() has gained an origin parameter¶
to_datetime() has gained a new parameter, origin, to define a reference date
from where to compute the resulting timestamps when parsing numerical values with a specific unit specified. (GH11276, GH11745)
For example, with 1960-01-01 as the starting date:
In [14]: pd.to_datetime([1, 2, 3], unit='D', origin=pd.Timestamp('1960-01-01'))
Out[14]: DatetimeIndex(['1960-01-02', '1960-01-03', '1960-01-04'], dtype='datetime64[ns]', freq=None)
The default is set at origin='unix', which defaults to 1970-01-01 00:00:00, which is
commonly called ‘unix epoch’ or POSIX time. This was the previous default, so this is a backward compatible change.
In [15]: pd.to_datetime([1, 2, 3], unit='D')
Out[15]: DatetimeIndex(['1970-01-02', '1970-01-03', '1970-01-04'], dtype='datetime64[ns]', freq=None)
Groupby Enhancements¶
Strings passed to DataFrame.groupby() as the by parameter may now reference either column names or index level names. Previously, only column names could be referenced. This allows to easily group by a column and index level at the same time. (GH5677)
In [16]: arrays = [['bar', 'bar', 'baz', 'baz', 'foo', 'foo', 'qux', 'qux'],
....: ['one', 'two', 'one', 'two', 'one', 'two', 'one', 'two']]
....:
In [17]: index = pd.MultiIndex.from_arrays(arrays, names=['first', 'second'])
In [18]: df = pd.DataFrame({'A': [1, 1, 1, 1, 2, 2, 3, 3],
....: 'B': np.arange(8)},
....: index=index)
....:
In [19]: df
Out[19]:
A B
first second
bar one 1 0
two 1 1
baz one 1 2
two 1 3
foo one 2 4
two 2 5
qux one 3 6
two 3 7
In [20]: df.groupby(['second', 'A']).sum()
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[20]:
B
second A
one 1 2
2 4
3 6
two 1 4
2 5
3 7
Better support for compressed URLs in read_csv¶
The compression code was refactored (GH12688). As a result, reading
dataframes from URLs in read_csv() or read_table() now supports
additional compression methods: xz, bz2, and zip (GH14570).
Previously, only gzip compression was supported. By default, compression of
URLs and paths are now inferred using their file extensions. Additionally,
support for bz2 compression in the python 2 C-engine improved (GH14874).
In [21]: url = 'https://github.com/{repo}/raw/{branch}/{path}'.format(
....: repo = 'pandas-dev/pandas',
....: branch = 'master',
....: path = 'pandas/tests/io/parser/data/salaries.csv.bz2',
....: )
....:
In [22]: df = pd.read_table(url, compression='infer') # default, infer compression
In [23]: df = pd.read_table(url, compression='bz2') # explicitly specify compression
In [24]: df.head(2)
Out[24]:
S X E M
0 13876 1 1 1
1 11608 1 3 0
Pickle file I/O now supports compression¶
read_pickle(), DataFrame.to_pickle() and Series.to_pickle()
can now read from and write to compressed pickle files. Compression methods
can be an explicit parameter or be inferred from the file extension.
See the docs here.
In [25]: df = pd.DataFrame({
....: 'A': np.random.randn(1000),
....: 'B': 'foo',
....: 'C': pd.date_range('20130101', periods=1000, freq='s')})
....:
Using an explicit compression type
In [26]: df.to_pickle("data.pkl.compress", compression="gzip")
In [27]: rt = pd.read_pickle("data.pkl.compress", compression="gzip")
In [28]: rt.head()
Out[28]:
A B C
0 0.384316 foo 2013-01-01 00:00:00
1 1.574159 foo 2013-01-01 00:00:01
2 1.588931 foo 2013-01-01 00:00:02
3 0.476720 foo 2013-01-01 00:00:03
4 0.473424 foo 2013-01-01 00:00:04
The default is to infer the compression type from the extension (compression='infer'):
In [29]: df.to_pickle("data.pkl.gz")
In [30]: rt = pd.read_pickle("data.pkl.gz")
In [31]: rt.head()
Out[31]:
A B C
0 0.384316 foo 2013-01-01 00:00:00
1 1.574159 foo 2013-01-01 00:00:01
2 1.588931 foo 2013-01-01 00:00:02
3 0.476720 foo 2013-01-01 00:00:03
4 0.473424 foo 2013-01-01 00:00:04
In [32]: df["A"].to_pickle("s1.pkl.bz2")
In [33]: rt = pd.read_pickle("s1.pkl.bz2")
In [34]: rt.head()
Out[34]:
0 0.384316
1 1.574159
2 1.588931
3 0.476720
4 0.473424
Name: A, dtype: float64
UInt64 Support Improved¶
Pandas has significantly improved support for operations involving unsigned,
or purely non-negative, integers. Previously, handling these integers would
result in improper rounding or data-type casting, leading to incorrect results.
Notably, a new numerical index, UInt64Index, has been created (GH14937)
In [35]: idx = pd.UInt64Index([1, 2, 3])
In [36]: df = pd.DataFrame({'A': ['a', 'b', 'c']}, index=idx)
In [37]: df.index
Out[37]: UInt64Index([1, 2, 3], dtype='uint64')
- Bug in converting object elements of array-like objects to unsigned 64-bit integers (GH4471, GH14982)
- Bug in
Series.unique()in which unsigned 64-bit integers were causing overflow (GH14721) - Bug in
DataFrameconstruction in which unsigned 64-bit integer elements were being converted to objects (GH14881) - Bug in
pd.read_csv()in which unsigned 64-bit integer elements were being improperly converted to the wrong data types (GH14983) - Bug in
pd.unique()in which unsigned 64-bit integers were causing overflow (GH14915) - Bug in
pd.value_counts()in which unsigned 64-bit integers were being erroneously truncated in the output (GH14934)
GroupBy on Categoricals¶
In previous versions, .groupby(..., sort=False) would fail with a ValueError when grouping on a categorical series with some categories not appearing in the data. (GH13179)
In [38]: chromosomes = np.r_[np.arange(1, 23).astype(str), ['X', 'Y']]
In [39]: df = pd.DataFrame({
....: 'A': np.random.randint(100),
....: 'B': np.random.randint(100),
....: 'C': np.random.randint(100),
....: 'chromosomes': pd.Categorical(np.random.choice(chromosomes, 100),
....: categories=chromosomes,
....: ordered=True)})
....:
In [40]: df
Out[40]:
A B C chromosomes
0 21 62 10 17
1 21 62 10 Y
2 21 62 10 13
3 21 62 10 8
4 21 62 10 22
5 21 62 10 3
6 21 62 10 19
.. .. .. .. ...
93 21 62 10 17
94 21 62 10 Y
95 21 62 10 Y
96 21 62 10 22
97 21 62 10 5
98 21 62 10 20
99 21 62 10 X
[100 rows x 4 columns]
Previous Behavior:
In [3]: df[df.chromosomes != '1'].groupby('chromosomes', sort=False).sum()
---------------------------------------------------------------------------
ValueError: items in new_categories are not the same as in old categories
New Behavior:
In [41]: df[df.chromosomes != '1'].groupby('chromosomes', sort=False).sum()
Out[41]:
A B C
chromosomes
2 42.0 124.0 20.0
3 105.0 310.0 50.0
4 63.0 186.0 30.0
5 84.0 248.0 40.0
6 84.0 248.0 40.0
7 63.0 186.0 30.0
8 189.0 558.0 90.0
... ... ... ...
20 126.0 372.0 60.0
21 42.0 124.0 20.0
22 84.0 248.0 40.0
X 63.0 186.0 30.0
Y 126.0 372.0 60.0
1 NaN NaN NaN
12 NaN NaN NaN
[24 rows x 3 columns]
Table Schema Output¶
The new orient 'table' for DataFrame.to_json()
will generate a Table Schema compatible string representation of
the data.
In [42]: df = pd.DataFrame(
....: {'A': [1, 2, 3],
....: 'B': ['a', 'b', 'c'],
....: 'C': pd.date_range('2016-01-01', freq='d', periods=3),
....: }, index=pd.Index(range(3), name='idx'))
....:
In [43]: df
Out[43]:
A B C
idx
0 1 a 2016-01-01
1 2 b 2016-01-02
2 3 c 2016-01-03
In [44]: df.to_json(orient='table')
�������������������������������������������������������������������������������������������������������������������Out[44]: '{"schema": {"fields":[{"name":"idx","type":"integer"},{"name":"A","type":"integer"},{"name":"B","type":"string"},{"name":"C","type":"datetime"}],"primaryKey":["idx"],"pandas_version":"0.20.0"}, "data": [{"idx":0,"A":1,"B":"a","C":"2016-01-01T00:00:00.000Z"},{"idx":1,"A":2,"B":"b","C":"2016-01-02T00:00:00.000Z"},{"idx":2,"A":3,"B":"c","C":"2016-01-03T00:00:00.000Z"}]}'
See IO: Table Schema for more information.
Additionally, the repr for DataFrame and Series can now publish
this JSON Table schema representation of the Series or DataFrame if you are
using IPython (or another frontend like nteract using the Jupyter messaging
protocol).
This gives frontends like the Jupyter notebook and nteract
more flexiblity in how they display pandas objects, since they have
more information about the data.
You must enable this by setting the display.html.table_schema option to True.
SciPy sparse matrix from/to SparseDataFrame¶
Pandas now supports creating sparse dataframes directly from scipy.sparse.spmatrix instances.
See the documentation for more information. (GH4343)
All sparse formats are supported, but matrices that are not in COOrdinate format will be converted, copying data as needed.
In [45]: from scipy.sparse import csr_matrix
In [46]: arr = np.random.random(size=(1000, 5))
In [47]: arr[arr < .9] = 0
In [48]: sp_arr = csr_matrix(arr)
In [49]: sp_arr
Out[49]:
<1000x5 sparse matrix of type '<class 'numpy.float64'>'
with 500 stored elements in Compressed Sparse Row format>
In [50]: sdf = pd.SparseDataFrame(sp_arr)
In [51]: sdf
Out[51]:
0 1 2 3 4
0 NaN NaN NaN NaN NaN
1 NaN NaN NaN NaN NaN
2 NaN NaN NaN NaN NaN
3 NaN NaN NaN NaN 0.997522
4 NaN NaN NaN NaN NaN
5 NaN NaN NaN NaN 0.911034
6 NaN NaN NaN NaN NaN
.. ... .. .. .. ...
993 0.925879 NaN NaN NaN NaN
994 NaN NaN NaN NaN 0.955585
995 NaN NaN NaN NaN NaN
996 NaN NaN NaN NaN NaN
997 NaN NaN NaN NaN NaN
998 NaN NaN NaN NaN 0.904855
999 NaN NaN NaN NaN NaN
[1000 rows x 5 columns]
To convert a SparseDataFrame back to sparse SciPy matrix in COO format, you can use:
In [52]: sdf.to_coo()
Out[52]:
<1000x5 sparse matrix of type '<class 'numpy.float64'>'
with 500 stored elements in COOrdinate format>
Excel output for styled DataFrames¶
Experimental support has been added to export DataFrame.style formats to Excel using the openpyxl engine. (GH15530)
For example, after running the following, styled.xlsx renders as below:
In [53]: np.random.seed(24)
In [54]: df = pd.DataFrame({'A': np.linspace(1, 10, 10)})
In [55]: df = pd.concat([df, pd.DataFrame(np.random.RandomState(24).randn(10, 4),
....: columns=list('BCDE'))],
....: axis=1)
....:
In [56]: df.iloc[0, 2] = np.nan
In [57]: df
Out[57]:
A B C D E
0 1.0 1.329212 NaN -0.316280 -0.990810
1 2.0 -1.070816 -1.438713 0.564417 0.295722
2 3.0 -1.626404 0.219565 0.678805 1.889273
3 4.0 0.961538 0.104011 -0.481165 0.850229
4 5.0 1.453425 1.057737 0.165562 0.515018
5 6.0 -1.336936 0.562861 1.392855 -0.063328
6 7.0 0.121668 1.207603 -0.002040 1.627796
7 8.0 0.354493 1.037528 -0.385684 0.519818
8 9.0 1.686583 -1.325963 1.428984 -2.089354
9 10.0 -0.129820 0.631523 -0.586538 0.290720
In [58]: styled = df.style.\
....: applymap(lambda val: 'color: %s' % 'red' if val < 0 else 'black').\
....: highlight_max()
....:
In [59]: styled.to_excel('styled.xlsx', engine='openpyxl')
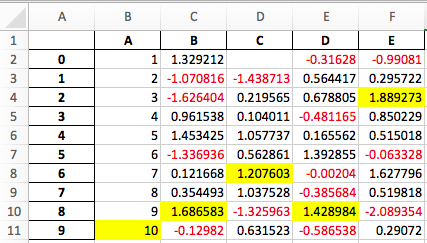
See the Style documentation for more detail.
IntervalIndex¶
pandas has gained an IntervalIndex with its own dtype, interval as well as the Interval scalar type. These allow first-class support for interval
notation, specifically as a return type for the categories in cut() and qcut(). The IntervalIndex allows some unique indexing, see the
docs. (GH7640, GH8625)
Warning
These indexing behaviors of the IntervalIndex are provisional and may change in a future version of pandas. Feedback on usage is welcome.
Previous behavior:
The returned categories were strings, representing Intervals
In [1]: c = pd.cut(range(4), bins=2)
In [2]: c
Out[2]:
[(-0.003, 1.5], (-0.003, 1.5], (1.5, 3], (1.5, 3]]
Categories (2, object): [(-0.003, 1.5] < (1.5, 3]]
In [3]: c.categories
Out[3]: Index(['(-0.003, 1.5]', '(1.5, 3]'], dtype='object')
New behavior:
In [60]: c = pd.cut(range(4), bins=2)
In [61]: c
Out[61]:
[(-0.003, 1.5], (-0.003, 1.5], (1.5, 3.0], (1.5, 3.0]]
Categories (2, interval[float64]): [(-0.003, 1.5] < (1.5, 3.0]]
In [62]: c.categories
���������������������������������������������������������������������������������������������������������������������������������Out[62]:
IntervalIndex([(-0.003, 1.5], (1.5, 3.0]]
closed='right',
dtype='interval[float64]')
Furthermore, this allows one to bin other data with these same bins, with NaN representing a missing
value similar to other dtypes.
In [63]: pd.cut([0, 3, 5, 1], bins=c.categories)
Out[63]:
[(-0.003, 1.5], (1.5, 3.0], NaN, (-0.003, 1.5]]
Categories (2, interval[float64]): [(-0.003, 1.5] < (1.5, 3.0]]
An IntervalIndex can also be used in Series and DataFrame as the index.
In [64]: df = pd.DataFrame({'A': range(4),
....: 'B': pd.cut([0, 3, 1, 1], bins=c.categories)}
....: ).set_index('B')
....:
In [65]: df
Out[65]:
A
B
(-0.003, 1.5] 0
(1.5, 3.0] 1
(-0.003, 1.5] 2
(-0.003, 1.5] 3
Selecting via a specific interval:
In [66]: df.loc[pd.Interval(1.5, 3.0)]
Out[66]:
A 1
Name: (1.5, 3.0], dtype: int64
Selecting via a scalar value that is contained in the intervals.
In [67]: df.loc[0]
Out[67]:
A
B
(-0.003, 1.5] 0
(-0.003, 1.5] 2
(-0.003, 1.5] 3
Other Enhancements¶
DataFrame.rolling()now accepts the parameterclosed='right'|'left'|'both'|'neither'to choose the rolling window-endpoint closedness. See the documentation (GH13965)- Integration with the
feather-format, including a new top-levelpd.read_feather()andDataFrame.to_feather()method, see here. Series.str.replace()now accepts a callable, as replacement, which is passed tore.sub(GH15055)Series.str.replace()now accepts a compiled regular expression as a pattern (GH15446)Series.sort_indexaccepts parameterskindandna_position(GH13589, GH14444)DataFrameandDataFrame.groupby()have gained anunique()method to count the distinct values over an axis (GH14336, GH15197).DataFramehas gained amelt()method, equivalent topd.melt(), for unpivoting from a wide to long format (GH12640).pd.read_excel()now preserves sheet order when usingsheetname=None(GH9930)- Multiple offset aliases with decimal points are now supported (e.g.
0.5minis parsed as30s) (GH8419) .isnull()and.notnull()have been added toIndexobject to make them more consistent with theSeriesAPI (GH15300)- New
UnsortedIndexError(subclass ofKeyError) raised when indexing/slicing into an unsorted MultiIndex (GH11897). This allows differentiation between errors due to lack of sorting or an incorrect key. See here MultiIndexhas gained a.to_frame()method to convert to aDataFrame(GH12397)pd.cutandpd.qcutnow support datetime64 and timedelta64 dtypes (GH14714, GH14798)pd.qcuthas gained theduplicates='raise'|'drop'option to control whether to raise on duplicated edges (GH7751)Seriesprovides ato_excelmethod to output Excel files (GH8825)- The
usecolsargument inpd.read_csv()now accepts a callable function as a value (GH14154) - The
skiprowsargument inpd.read_csv()now accepts a callable function as a value (GH10882) - The
nrowsandchunksizearguments inpd.read_csv()are supported if both are passed (GH6774, GH15755) DataFrame.plotnow prints a title above each subplot ifsuplots=Trueandtitleis a list of strings (GH14753)DataFrame.plotcan pass the matplotlib 2.0 default color cycle as a single string as color parameter, see here. (GH15516)Series.interpolate()now supports timedelta as an index type withmethod='time'(GH6424)- Addition of a
levelkeyword toDataFrame/Series.renameto rename labels in the specified level of a MultiIndex (GH4160). DataFrame.reset_index()will now interpret a tupleindex.nameas a key spanning across levels ofcolumns, if this is aMultiIndex(GH16164)Timedelta.isoformatmethod added for formatting Timedeltas as an ISO 8601 duration. See the Timedelta docs (GH15136).select_dtypes()now allows the stringdatetimetzto generically select datetimes with tz (GH14910)- The
.to_latex()method will now acceptmulticolumnandmultirowarguments to use the accompanying LaTeX enhancements pd.merge_asof()gained the optiondirection='backward'|'forward'|'nearest'(GH14887)Series/DataFrame.asfreq()have gained afill_valueparameter, to fill missing values (GH3715).Series/DataFrame.resample.asfreqhave gained afill_valueparameter, to fill missing values during resampling (GH3715).pandas.util.hash_pandas_object()has gained the ability to hash aMultiIndex(GH15224)Series/DataFrame.squeeze()have gained theaxisparameter. (GH15339)DataFrame.to_excel()has a newfreeze_panesparameter to turn on Freeze Panes when exporting to Excel (GH15160)pd.read_html()will parse multiple header rows, creating a MutliIndex header. (GH13434).- HTML table output skips
colspanorrowspanattribute if equal to 1. (GH15403) pandas.io.formats.style.Stylertemplate now has blocks for easier extension, see the example notebook (GH15649)Styler.render()now accepts**kwargsto allow user-defined variables in the template (GH15649)- Compatibility with Jupyter notebook 5.0; MultiIndex column labels are left-aligned and MultiIndex row-labels are top-aligned (GH15379)
TimedeltaIndexnow has a custom date-tick formatter specifically designed for nanosecond level precision (GH8711)pd.api.types.union_categoricalsgained theignore_orderedargument to allow ignoring the ordered attribute of unioned categoricals (GH13410). See the categorical union docs for more information.DataFrame.to_latex()andDataFrame.to_string()now allow optional header aliases. (GH15536)- Re-enable the
parse_dateskeyword ofpd.read_excel()to parse string columns as dates (GH14326) - Added
.emptyproperty to subclasses ofIndex. (GH15270) - Enabled floor division for
TimedeltaandTimedeltaIndex(GH15828) pandas.io.json.json_normalize()gained the optionerrors='ignore'|'raise'; the default iserrors='raise'which is backward compatible. (GH14583)pandas.io.json.json_normalize()with an emptylistwill return an emptyDataFrame(GH15534)pandas.io.json.json_normalize()has gained asepoption that acceptsstrto separate joined fields; the default is ”.”, which is backward compatible. (GH14883)MultiIndex.remove_unused_levels()has been added to facilitate removing unused levels. (GH15694)pd.read_csv()will now raise aParserErrorerror whenever any parsing error occurs (GH15913, GH15925)pd.read_csv()now supports theerror_bad_linesandwarn_bad_linesarguments for the Python parser (GH15925)- The
display.show_dimensionsoption can now also be used to specify whether the length of aSeriesshould be shown in its repr (GH7117). parallel_coordinates()has gained asort_labelskeyword argument that sorts class labels and the colors assigned to them (GH15908)- Options added to allow one to turn on/off using
bottleneckandnumexpr, see here (GH16157) DataFrame.style.bar()now accepts two more options to further customize the bar chart. Bar alignment is set withalign='left'|'mid'|'zero', the default is “left”, which is backward compatible; You can now pass a list ofcolor=[color_negative, color_positive]. (GH14757)
Backwards incompatible API changes¶
Possible incompatibility for HDF5 formats created with pandas < 0.13.0¶
pd.TimeSeries was deprecated officially in 0.17.0, though has already been an alias since 0.13.0. It has
been dropped in favor of pd.Series. (GH15098).
This may cause HDF5 files that were created in prior versions to become unreadable if pd.TimeSeries
was used. This is most likely to be for pandas < 0.13.0. If you find yourself in this situation.
You can use a recent prior version of pandas to read in your HDF5 files,
then write them out again after applying the procedure below.
In [2]: s = pd.TimeSeries([1,2,3], index=pd.date_range('20130101', periods=3))
In [3]: s
Out[3]:
2013-01-01 1
2013-01-02 2
2013-01-03 3
Freq: D, dtype: int64
In [4]: type(s)
Out[4]: pandas.core.series.TimeSeries
In [5]: s = pd.Series(s)
In [6]: s
Out[6]:
2013-01-01 1
2013-01-02 2
2013-01-03 3
Freq: D, dtype: int64
In [7]: type(s)
Out[7]: pandas.core.series.Series
Map on Index types now return other Index types¶
map on an Index now returns an Index, not a numpy array (GH12766)
In [68]: idx = Index([1, 2])
In [69]: idx
Out[69]: Int64Index([1, 2], dtype='int64')
In [70]: mi = MultiIndex.from_tuples([(1, 2), (2, 4)])
In [71]: mi
Out[71]:
MultiIndex(levels=[[1, 2], [2, 4]],
labels=[[0, 1], [0, 1]])
Previous Behavior:
In [5]: idx.map(lambda x: x * 2)
Out[5]: array([2, 4])
In [6]: idx.map(lambda x: (x, x * 2))
Out[6]: array([(1, 2), (2, 4)], dtype=object)
In [7]: mi.map(lambda x: x)
Out[7]: array([(1, 2), (2, 4)], dtype=object)
In [8]: mi.map(lambda x: x[0])
Out[8]: array([1, 2])
New Behavior:
In [72]: idx.map(lambda x: x * 2)
Out[72]: Int64Index([2, 4], dtype='int64')
In [73]: idx.map(lambda x: (x, x * 2))
�������������������������������������������Out[73]:
MultiIndex(levels=[[1, 2], [2, 4]],
labels=[[0, 1], [0, 1]])
In [74]: mi.map(lambda x: x)
�����������������������������������������������������������������������������������������������������������������������������Out[74]:
MultiIndex(levels=[[1, 2], [2, 4]],
labels=[[0, 1], [0, 1]])
In [75]: mi.map(lambda x: x[0])
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[75]: Int64Index([1, 2], dtype='int64')
map on a Series with datetime64 values may return int64 dtypes rather than int32
In [76]: s = Series(date_range('2011-01-02T00:00', '2011-01-02T02:00', freq='H').tz_localize('Asia/Tokyo'))
In [77]: s
Out[77]:
0 2011-01-02 00:00:00+09:00
1 2011-01-02 01:00:00+09:00
2 2011-01-02 02:00:00+09:00
dtype: datetime64[ns, Asia/Tokyo]
Previous Behavior:
In [9]: s.map(lambda x: x.hour)
Out[9]:
0 0
1 1
2 2
dtype: int32
New Behavior:
In [78]: s.map(lambda x: x.hour)
Out[78]:
0 0
1 1
2 2
dtype: int64
Accessing datetime fields of Index now return Index¶
The datetime-related attributes (see here
for an overview) of DatetimeIndex, PeriodIndex and TimedeltaIndex previously
returned numpy arrays. They will now return a new Index object, except
in the case of a boolean field, where the result will still be a boolean ndarray. (GH15022)
Previous behaviour:
In [1]: idx = pd.date_range("2015-01-01", periods=5, freq='10H')
In [2]: idx.hour
Out[2]: array([ 0, 10, 20, 6, 16], dtype=int32)
New Behavior:
In [79]: idx = pd.date_range("2015-01-01", periods=5, freq='10H')
In [80]: idx.hour
Out[80]: Int64Index([0, 10, 20, 6, 16], dtype='int64')
This has the advantage that specific Index methods are still available on the
result. On the other hand, this might have backward incompatibilities: e.g.
compared to numpy arrays, Index objects are not mutable. To get the original
ndarray, you can always convert explicitly using np.asarray(idx.hour).
pd.unique will now be consistent with extension types¶
In prior versions, using Series.unique() and pandas.unique() on Categorical and tz-aware
data-types would yield different return types. These are now made consistent. (GH15903)
Datetime tz-aware
Previous behaviour:
# Series In [5]: pd.Series([pd.Timestamp('20160101', tz='US/Eastern'), pd.Timestamp('20160101', tz='US/Eastern')]).unique() Out[5]: array([Timestamp('2016-01-01 00:00:00-0500', tz='US/Eastern')], dtype=object) In [6]: pd.unique(pd.Series([pd.Timestamp('20160101', tz='US/Eastern'), pd.Timestamp('20160101', tz='US/Eastern')])) Out[6]: array(['2016-01-01T05:00:00.000000000'], dtype='datetime64[ns]') # Index In [7]: pd.Index([pd.Timestamp('20160101', tz='US/Eastern'), pd.Timestamp('20160101', tz='US/Eastern')]).unique() Out[7]: DatetimeIndex(['2016-01-01 00:00:00-05:00'], dtype='datetime64[ns, US/Eastern]', freq=None) In [8]: pd.unique([pd.Timestamp('20160101', tz='US/Eastern'), pd.Timestamp('20160101', tz='US/Eastern')]) Out[8]: array(['2016-01-01T05:00:00.000000000'], dtype='datetime64[ns]')
New Behavior:
# Series, returns an array of Timestamp tz-aware In [81]: pd.Series([pd.Timestamp('20160101', tz='US/Eastern'), ....: pd.Timestamp('20160101', tz='US/Eastern')]).unique() ....: Out[81]: array([Timestamp('2016-01-01 00:00:00-0500', tz='US/Eastern')], dtype=object) In [82]: pd.unique(pd.Series([pd.Timestamp('20160101', tz='US/Eastern'), ....: pd.Timestamp('20160101', tz='US/Eastern')])) ....: ���������������������������������������������������������������������������������������Out[82]: array([Timestamp('2016-01-01 00:00:00-0500', tz='US/Eastern')], dtype=object) # Index, returns a DatetimeIndex In [83]: pd.Index([pd.Timestamp('20160101', tz='US/Eastern'), ....: pd.Timestamp('20160101', tz='US/Eastern')]).unique() ....: ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[83]: DatetimeIndex(['2016-01-01 00:00:00-05:00'], dtype='datetime64[ns, US/Eastern]', freq=None) In [84]: pd.unique(pd.Index([pd.Timestamp('20160101', tz='US/Eastern'), ....: pd.Timestamp('20160101', tz='US/Eastern')])) ....: �����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[84]: DatetimeIndex(['2016-01-01 00:00:00-05:00'], dtype='datetime64[ns, US/Eastern]', freq=None)
Categoricals
Previous behaviour:
In [1]: pd.Series(list('baabc'), dtype='category').unique() Out[1]: [b, a, c] Categories (3, object): [b, a, c] In [2]: pd.unique(pd.Series(list('baabc'), dtype='category')) Out[2]: array(['b', 'a', 'c'], dtype=object)
New Behavior:
# returns a Categorical In [85]: pd.Series(list('baabc'), dtype='category').unique() Out[85]: [b, a, c] Categories (3, object): [b, a, c] In [86]: pd.unique(pd.Series(list('baabc'), dtype='category')) ������������������������������������������������������Out[86]: [b, a, c] Categories (3, object): [b, a, c]
S3 File Handling¶
pandas now uses s3fs for handling S3 connections. This shouldn’t break
any code. However, since s3fs is not a required dependency, you will need to install it separately, like boto
in prior versions of pandas. (GH11915).
Partial String Indexing Changes¶
DatetimeIndex Partial String Indexing now works as an exact match, provided that string resolution coincides with index resolution, including a case when both are seconds (GH14826). See Slice vs. Exact Match for details.
In [87]: df = DataFrame({'a': [1, 2, 3]}, DatetimeIndex(['2011-12-31 23:59:59',
....: '2012-01-01 00:00:00',
....: '2012-01-01 00:00:01']))
....:
Previous Behavior:
In [4]: df['2011-12-31 23:59:59']
Out[4]:
a
2011-12-31 23:59:59 1
In [5]: df['a']['2011-12-31 23:59:59']
Out[5]:
2011-12-31 23:59:59 1
Name: a, dtype: int64
New Behavior:
In [4]: df['2011-12-31 23:59:59']
KeyError: '2011-12-31 23:59:59'
In [5]: df['a']['2011-12-31 23:59:59']
Out[5]: 1
Concat of different float dtypes will not automatically upcast¶
Previously, concat of multiple objects with different float dtypes would automatically upcast results to a dtype of float64.
Now the smallest acceptable dtype will be used (GH13247)
In [88]: df1 = pd.DataFrame(np.array([1.0], dtype=np.float32, ndmin=2))
In [89]: df1.dtypes
Out[89]:
0 float32
dtype: object
In [90]: df2 = pd.DataFrame(np.array([np.nan], dtype=np.float32, ndmin=2))
In [91]: df2.dtypes
Out[91]:
0 float32
dtype: object
Previous Behavior:
In [7]: pd.concat([df1, df2]).dtypes
Out[7]:
0 float64
dtype: object
New Behavior:
In [92]: pd.concat([df1, df2]).dtypes
Out[92]:
0 float32
dtype: object
Pandas Google BigQuery support has moved¶
pandas has split off Google BigQuery support into a separate package pandas-gbq. You can conda install pandas-gbq -c conda-forge or
pip install pandas-gbq to get it. The functionality of read_gbq() and DataFrame.to_gbq() remain the same with the
currently released version of pandas-gbq=0.1.4. Documentation is now hosted here (GH15347)
Memory Usage for Index is more Accurate¶
In previous versions, showing .memory_usage() on a pandas structure that has an index, would only include actual index values and not include structures that facilitated fast indexing. This will generally be different for Index and MultiIndex and less-so for other index types. (GH15237)
Previous Behavior:
In [8]: index = Index(['foo', 'bar', 'baz'])
In [9]: index.memory_usage(deep=True)
Out[9]: 180
In [10]: index.get_loc('foo')
Out[10]: 0
In [11]: index.memory_usage(deep=True)
Out[11]: 180
New Behavior:
In [8]: index = Index(['foo', 'bar', 'baz'])
In [9]: index.memory_usage(deep=True)
Out[9]: 180
In [10]: index.get_loc('foo')
Out[10]: 0
In [11]: index.memory_usage(deep=True)
Out[11]: 260
DataFrame.sort_index changes¶
In certain cases, calling .sort_index() on a MultiIndexed DataFrame would return the same DataFrame without seeming to sort.
This would happen with a lexsorted, but non-monotonic levels. (GH15622, GH15687, GH14015, GH13431, GH15797)
This is unchanged from prior versions, but shown for illustration purposes:
In [93]: df = DataFrame(np.arange(6), columns=['value'], index=MultiIndex.from_product([list('BA'), range(3)]))
In [94]: df
Out[94]:
value
B 0 0
1 1
2 2
A 0 3
1 4
2 5
In [95]: df.index.is_lexsorted()
Out[95]: False
In [96]: df.index.is_monotonic
���������������Out[96]: False
Sorting works as expected
In [97]: df.sort_index()
Out[97]:
value
A 0 3
1 4
2 5
B 0 0
1 1
2 2
In [98]: df.sort_index().index.is_lexsorted()
Out[98]: True
In [99]: df.sort_index().index.is_monotonic
��������������Out[99]: True
However, this example, which has a non-monotonic 2nd level, doesn’t behave as desired.
In [100]: df = pd.DataFrame(
.....: {'value': [1, 2, 3, 4]},
.....: index=pd.MultiIndex(levels=[['a', 'b'], ['bb', 'aa']],
.....: labels=[[0, 0, 1, 1], [0, 1, 0, 1]]))
.....:
In [101]: df
Out[101]:
value
a bb 1
aa 2
b bb 3
aa 4
Previous Behavior:
In [11]: df.sort_index()
Out[11]:
value
a bb 1
aa 2
b bb 3
aa 4
In [14]: df.sort_index().index.is_lexsorted()
Out[14]: True
In [15]: df.sort_index().index.is_monotonic
Out[15]: False
New Behavior:
In [102]: df.sort_index()
Out[102]:
value
a aa 2
bb 1
b aa 4
bb 3
In [103]: df.sort_index().index.is_lexsorted()
�����������������������������������������������������������������������Out[103]: True
In [104]: df.sort_index().index.is_monotonic
��������������������������������������������������������������������������������������Out[104]: True
Groupby Describe Formatting¶
The output formatting of groupby.describe() now labels the describe() metrics in the columns instead of the index.
This format is consistent with groupby.agg() when applying multiple functions at once. (GH4792)
Previous Behavior:
In [1]: df = pd.DataFrame({'A': [1, 1, 2, 2], 'B': [1, 2, 3, 4]})
In [2]: df.groupby('A').describe()
Out[2]:
B
A
1 count 2.000000
mean 1.500000
std 0.707107
min 1.000000
25% 1.250000
50% 1.500000
75% 1.750000
max 2.000000
2 count 2.000000
mean 3.500000
std 0.707107
min 3.000000
25% 3.250000
50% 3.500000
75% 3.750000
max 4.000000
In [3]: df.groupby('A').agg([np.mean, np.std, np.min, np.max])
Out[3]:
B
mean std amin amax
A
1 1.5 0.707107 1 2
2 3.5 0.707107 3 4
New Behavior:
In [105]: df = pd.DataFrame({'A': [1, 1, 2, 2], 'B': [1, 2, 3, 4]})
In [106]: df.groupby('A').describe()
Out[106]:
B
count mean std min 25% 50% 75% max
A
1 2.0 1.5 0.707107 1.0 1.25 1.5 1.75 2.0
2 2.0 3.5 0.707107 3.0 3.25 3.5 3.75 4.0
In [107]: df.groupby('A').agg([np.mean, np.std, np.min, np.max])
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[107]:
B
mean std amin amax
A
1 1.5 0.707107 1 2
2 3.5 0.707107 3 4
Window Binary Corr/Cov operations return a MultiIndex DataFrame¶
A binary window operation, like .corr() or .cov(), when operating on a .rolling(..), .expanding(..), or .ewm(..) object,
will now return a 2-level MultiIndexed DataFrame rather than a Panel, as Panel is now deprecated,
see here. These are equivalent in function,
but a MultiIndexed DataFrame enjoys more support in pandas.
See the section on Windowed Binary Operations for more information. (GH15677)
In [108]: np.random.seed(1234)
In [109]: df = pd.DataFrame(np.random.rand(100, 2),
.....: columns=pd.Index(['A', 'B'], name='bar'),
.....: index=pd.date_range('20160101',
.....: periods=100, freq='D', name='foo'))
.....:
In [110]: df.tail()
Out[110]:
bar A B
foo
2016-04-05 0.640880 0.126205
2016-04-06 0.171465 0.737086
2016-04-07 0.127029 0.369650
2016-04-08 0.604334 0.103104
2016-04-09 0.802374 0.945553
Previous Behavior:
In [2]: df.rolling(12).corr()
Out[2]:
<class 'pandas.core.panel.Panel'>
Dimensions: 100 (items) x 2 (major_axis) x 2 (minor_axis)
Items axis: 2016-01-01 00:00:00 to 2016-04-09 00:00:00
Major_axis axis: A to B
Minor_axis axis: A to B
New Behavior:
In [111]: res = df.rolling(12).corr()
In [112]: res.tail()
Out[112]:
bar A B
foo bar
2016-04-07 B -0.132090 1.000000
2016-04-08 A 1.000000 -0.145775
B -0.145775 1.000000
2016-04-09 A 1.000000 0.119645
B 0.119645 1.000000
Retrieving a correlation matrix for a cross-section
In [113]: df.rolling(12).corr().loc['2016-04-07']
Out[113]:
bar A B
foo bar
2016-04-07 A 1.00000 -0.13209
B -0.13209 1.00000
HDFStore where string comparison¶
In previous versions most types could be compared to string column in a HDFStore
usually resulting in an invalid comparison, returning an empty result frame. These comparisons will now raise a
TypeError (GH15492)
In [114]: df = pd.DataFrame({'unparsed_date': ['2014-01-01', '2014-01-01']})
In [115]: df.to_hdf('store.h5', 'key', format='table', data_columns=True)
In [116]: df.dtypes
Out[116]:
unparsed_date object
dtype: object
Previous Behavior:
In [4]: pd.read_hdf('store.h5', 'key', where='unparsed_date > ts')
File "<string>", line 1
(unparsed_date > 1970-01-01 00:00:01.388552400)
^
SyntaxError: invalid token
New Behavior:
In [18]: ts = pd.Timestamp('2014-01-01')
In [19]: pd.read_hdf('store.h5', 'key', where='unparsed_date > ts')
TypeError: Cannot compare 2014-01-01 00:00:00 of
type <class 'pandas.tslib.Timestamp'> to string column
Index.intersection and inner join now preserve the order of the left Index¶
Index.intersection() now preserves the order of the calling Index (left)
instead of the other Index (right) (GH15582). This affects inner
joins, DataFrame.join() and merge(), and the .align method.
Index.intersectionIn [117]: left = pd.Index([2, 1, 0]) In [118]: left Out[118]: Int64Index([2, 1, 0], dtype='int64') In [119]: right = pd.Index([1, 2, 3]) In [120]: right Out[120]: Int64Index([1, 2, 3], dtype='int64')
Previous Behavior:
In [4]: left.intersection(right) Out[4]: Int64Index([1, 2], dtype='int64')
New Behavior:
In [121]: left.intersection(right) Out[121]: Int64Index([2, 1], dtype='int64')
DataFrame.joinandpd.mergeIn [122]: left = pd.DataFrame({'a': [20, 10, 0]}, index=[2, 1, 0]) In [123]: left Out[123]: a 2 20 1 10 0 0 In [124]: right = pd.DataFrame({'b': [100, 200, 300]}, index=[1, 2, 3]) In [125]: right Out[125]: b 1 100 2 200 3 300
Previous Behavior:
In [4]: left.join(right, how='inner') Out[4]: a b 1 10 100 2 20 200
New Behavior:
In [126]: left.join(right, how='inner') Out[126]: a b 2 20 200 1 10 100
Pivot Table always returns a DataFrame¶
The documentation for pivot_table() states that a DataFrame is always returned. Here a bug
is fixed that allowed this to return a Series under certain circumstance. (GH4386)
In [127]: df = DataFrame({'col1': [3, 4, 5],
.....: 'col2': ['C', 'D', 'E'],
.....: 'col3': [1, 3, 9]})
.....:
In [128]: df
Out[128]:
col1 col2 col3
0 3 C 1
1 4 D 3
2 5 E 9
Previous Behavior:
In [2]: df.pivot_table('col1', index=['col3', 'col2'], aggfunc=np.sum)
Out[2]:
col3 col2
1 C 3
3 D 4
9 E 5
Name: col1, dtype: int64
New Behavior:
In [129]: df.pivot_table('col1', index=['col3', 'col2'], aggfunc=np.sum)
Out[129]:
col1
col3 col2
1 C 3
3 D 4
9 E 5
Other API Changes¶
numexprversion is now required to be >= 2.4.6 and it will not be used at all if this requisite is not fulfilled (GH15213).CParserErrorhas been renamed toParserErrorinpd.read_csv()and will be removed in the future (GH12665)SparseArray.cumsum()andSparseSeries.cumsum()will now always returnSparseArrayandSparseSeriesrespectively (GH12855)DataFrame.applymap()with an emptyDataFramewill return a copy of the emptyDataFrameinstead of aSeries(GH8222)Series.map()now respects default values of dictionary subclasses with a__missing__method, such ascollections.Counter(GH15999).lochas compat with.ixfor accepting iterators, and NamedTuples (GH15120)interpolate()andfillna()will raise aValueErrorif thelimitkeyword argument is not greater than 0. (GH9217)pd.read_csv()will now issue aParserWarningwhenever there are conflicting values provided by thedialectparameter and the user (GH14898)pd.read_csv()will now raise aValueErrorfor the C engine if the quote character is larger than than one byte (GH11592)inplacearguments now require a boolean value, else aValueErroris thrown (GH14189)pandas.api.types.is_datetime64_ns_dtypewill now reportTrueon a tz-aware dtype, similar topandas.api.types.is_datetime64_any_dtypeDataFrame.asof()will return a null filledSeriesinstead the scalarNaNif a match is not found (GH15118)- Specific support for
copy.copy()andcopy.deepcopy()functions on NDFrame objects (GH15444) Series.sort_values()accepts a one element list of bool for consistency with the behavior ofDataFrame.sort_values()(GH15604).merge()and.join()oncategorydtype columns will now preserve the category dtype when possible (GH10409)SparseDataFrame.default_fill_valuewill be 0, previously wasnanin the return frompd.get_dummies(..., sparse=True)(GH15594)- The default behaviour of
Series.str.matchhas changed from extracting groups to matching the pattern. The extracting behaviour was deprecated since pandas version 0.13.0 and can be done with theSeries.str.extractmethod (GH5224). As a consequence, theas_indexerkeyword is ignored (no longer needed to specify the new behaviour) and is deprecated. NaTwill now correctly reportFalsefor datetimelike boolean operations such asis_month_start(GH15781)NaTwill now correctly returnnp.nanforTimedeltaandPeriodaccessors such asdaysandquarter(GH15782)NaTwill now returnsNaTfortz_localizeandtz_convertmethods (GH15830)DataFrameandPanelconstructors with invalid input will now raiseValueErrorrather thanPandasError, if called with scalar inputs and not axes (GH15541)DataFrameandPanelconstructors with invalid input will now raiseValueErrorrather thanpandas.core.common.PandasError, if called with scalar inputs and not axes; The exceptionPandasErroris removed as well. (GH15541)- The exception
pandas.core.common.AmbiguousIndexErroris removed as it is not referenced (GH15541)
Reorganization of the library: Privacy Changes¶
Modules Privacy Has Changed¶
Some formerly public python/c/c++/cython extension modules have been moved and/or renamed. These are all removed from the public API.
Furthermore, the pandas.core, pandas.compat, and pandas.util top-level modules are now considered to be PRIVATE.
If indicated, a deprecation warning will be issued if you reference theses modules. (GH12588)
| Previous Location | New Location | Deprecated |
|---|---|---|
| pandas.lib | pandas._libs.lib | X |
| pandas.tslib | pandas._libs.tslib | X |
| pandas.computation | pandas.core.computation | X |
| pandas.msgpack | pandas.io.msgpack | |
| pandas.index | pandas._libs.index | |
| pandas.algos | pandas._libs.algos | |
| pandas.hashtable | pandas._libs.hashtable | |
| pandas.indexes | pandas.core.indexes | |
| pandas.json | pandas._libs.json / pandas.io.json | X |
| pandas.parser | pandas._libs.parsers | X |
| pandas.formats | pandas.io.formats | |
| pandas.sparse | pandas.core.sparse | |
| pandas.tools | pandas.core.reshape | X |
| pandas.types | pandas.core.dtypes | X |
| pandas.io.sas.saslib | pandas.io.sas._sas | |
| pandas._join | pandas._libs.join | |
| pandas._hash | pandas._libs.hashing | |
| pandas._period | pandas._libs.period | |
| pandas._sparse | pandas._libs.sparse | |
| pandas._testing | pandas._libs.testing | |
| pandas._window | pandas._libs.window |
Some new subpackages are created with public functionality that is not directly
exposed in the top-level namespace: pandas.errors, pandas.plotting and
pandas.testing (more details below). Together with pandas.api.types and
certain functions in the pandas.io and pandas.tseries submodules,
these are now the public subpackages.
Further changes:
- The function
union_categoricals()is now importable frompandas.api.types, formerly frompandas.types.concat(GH15998) - The type import
pandas.tslib.NaTTypeis deprecated and can be replaced by usingtype(pandas.NaT)(GH16146) - The public functions in
pandas.tools.hashingdeprecated from that locations, but are now importable frompandas.util(GH16223) - The modules in
pandas.util:decorators,print_versions,doctools,validators,depr_moduleare now private. Only the functions exposed inpandas.utilitself are public (GH16223)
pandas.errors¶
We are adding a standard public module for all pandas exceptions & warnings pandas.errors. (GH14800). Previously
these exceptions & warnings could be imported from pandas.core.common or pandas.io.common. These exceptions and warnings
will be removed from the *.common locations in a future release. (GH15541)
The following are now part of this API:
['DtypeWarning',
'EmptyDataError',
'OutOfBoundsDatetime',
'ParserError',
'ParserWarning',
'PerformanceWarning',
'UnsortedIndexError',
'UnsupportedFunctionCall']
pandas.testing¶
We are adding a standard module that exposes the public testing functions in pandas.testing (GH9895). Those functions can be used when writing tests for functionality using pandas objects.
The following testing functions are now part of this API:
pandas.plotting¶
A new public pandas.plotting module has been added that holds plotting functionality that was previously in either pandas.tools.plotting or in the top-level namespace. See the deprecations sections for more details.
Other Development Changes¶
Deprecations¶
Deprecate .ix¶
The .ix indexer is deprecated, in favor of the more strict .iloc and .loc indexers. .ix offers a lot of magic on the inference of what the user wants to do. To wit, .ix can decide to index positionally OR via labels, depending on the data type of the index. This has caused quite a bit of user confusion over the years. The full indexing documentation is here. (GH14218)
The recommended methods of indexing are:
.locif you want to label index.ilocif you want to positionally index.
Using .ix will now show a DeprecationWarning with a link to some examples of how to convert code here.
In [130]: df = pd.DataFrame({'A': [1, 2, 3],
.....: 'B': [4, 5, 6]},
.....: index=list('abc'))
.....:
In [131]: df
Out[131]:
A B
a 1 4
b 2 5
c 3 6
Previous Behavior, where you wish to get the 0th and the 2nd elements from the index in the ‘A’ column.
In [3]: df.ix[[0, 2], 'A']
Out[3]:
a 1
c 3
Name: A, dtype: int64
Using .loc. Here we will select the appropriate indexes from the index, then use label indexing.
In [132]: df.loc[df.index[[0, 2]], 'A']
Out[132]:
a 1
c 3
Name: A, dtype: int64
Using .iloc. Here we will get the location of the ‘A’ column, then use positional indexing to select things.
In [133]: df.iloc[[0, 2], df.columns.get_loc('A')]
Out[133]:
a 1
c 3
Name: A, dtype: int64
Deprecate Panel¶
Panel is deprecated and will be removed in a future version. The recommended way to represent 3-D data are
with a MultiIndex on a DataFrame via the to_frame() or with the xarray package. Pandas
provides a to_xarray() method to automate this conversion. For more details see Deprecate Panel documentation. (GH13563).
In [134]: p = tm.makePanel()
In [135]: p
Out[135]:
<class 'pandas.core.panel.Panel'>
Dimensions: 3 (items) x 3 (major_axis) x 4 (minor_axis)
Items axis: ItemA to ItemC
Major_axis axis: 2000-01-03 00:00:00 to 2000-01-05 00:00:00
Minor_axis axis: A to D
Convert to a MultiIndex DataFrame
In [136]: p.to_frame()
Out[136]:
ItemA ItemB ItemC
major minor
2000-01-03 A 0.628776 -1.409432 0.209395
B 0.988138 -1.347533 -0.896581
C -0.938153 1.272395 -0.161137
D -0.223019 -0.591863 -1.051539
2000-01-04 A 0.186494 1.422986 -0.592886
B -0.072608 0.363565 1.104352
C -1.239072 -1.449567 0.889157
D 2.123692 -0.414505 -0.319561
2000-01-05 A 0.952478 -2.147855 -1.473116
B -0.550603 -0.014752 -0.431550
C 0.139683 -1.195524 0.288377
D 0.122273 -1.425795 -0.619993
Convert to an xarray DataArray
In [137]: p.to_xarray()
Out[137]:
<xarray.DataArray (items: 3, major_axis: 3, minor_axis: 4)>
array([[[ 0.628776, 0.988138, -0.938153, -0.223019],
[ 0.186494, -0.072608, -1.239072, 2.123692],
[ 0.952478, -0.550603, 0.139683, 0.122273]],
[[-1.409432, -1.347533, 1.272395, -0.591863],
[ 1.422986, 0.363565, -1.449567, -0.414505],
[-2.147855, -0.014752, -1.195524, -1.425795]],
[[ 0.209395, -0.896581, -0.161137, -1.051539],
[-0.592886, 1.104352, 0.889157, -0.319561],
[-1.473116, -0.43155 , 0.288377, -0.619993]]])
Coordinates:
* items (items) object 'ItemA' 'ItemB' 'ItemC'
* major_axis (major_axis) datetime64[ns] 2000-01-03 2000-01-04 2000-01-05
* minor_axis (minor_axis) object 'A' 'B' 'C' 'D'
Deprecate groupby.agg() with a dictionary when renaming¶
The .groupby(..).agg(..), .rolling(..).agg(..), and .resample(..).agg(..) syntax can accept a variable of inputs, including scalars,
list, and a dict of column names to scalars or lists. This provides a useful syntax for constructing multiple
(potentially different) aggregations.
However, .agg(..) can also accept a dict that allows ‘renaming’ of the result columns. This is a complicated and confusing syntax, as well as not consistent
between Series and DataFrame. We are deprecating this ‘renaming’ functionaility.
- We are deprecating passing a dict to a grouped/rolled/resampled
Series. This allowed one torenamethe resulting aggregation, but this had a completely different meaning than passing a dictionary to a groupedDataFrame, which accepts column-to-aggregations. - We are deprecating passing a dict-of-dicts to a grouped/rolled/resampled
DataFramein a similar manner.
This is an illustrative example:
In [138]: df = pd.DataFrame({'A': [1, 1, 1, 2, 2],
.....: 'B': range(5),
.....: 'C': range(5)})
.....:
In [139]: df
Out[139]:
A B C
0 1 0 0
1 1 1 1
2 1 2 2
3 2 3 3
4 2 4 4
Here is a typical useful syntax for computing different aggregations for different columns. This
is a natural, and useful syntax. We aggregate from the dict-to-list by taking the specified
columns and applying the list of functions. This returns a MultiIndex for the columns (this is not deprecated).
In [140]: df.groupby('A').agg({'B': 'sum', 'C': 'min'})
Out[140]:
B C
A
1 3 0
2 7 3
Here’s an example of the first deprecation, passing a dict to a grouped Series. This
is a combination aggregation & renaming:
In [6]: df.groupby('A').B.agg({'foo': 'count'})
FutureWarning: using a dict on a Series for aggregation
is deprecated and will be removed in a future version
Out[6]:
foo
A
1 3
2 2
You can accomplish the same operation, more idiomatically by:
In [141]: df.groupby('A').B.agg(['count']).rename(columns={'count': 'foo'})
Out[141]:
foo
A
1 3
2 2
Here’s an example of the second deprecation, passing a dict-of-dict to a grouped DataFrame:
In [23]: (df.groupby('A')
.agg({'B': {'foo': 'sum'}, 'C': {'bar': 'min'}})
)
FutureWarning: using a dict with renaming is deprecated and
will be removed in a future version
Out[23]:
B C
foo bar
A
1 3 0
2 7 3
You can accomplish nearly the same by:
In [142]: (df.groupby('A')
.....: .agg({'B': 'sum', 'C': 'min'})
.....: .rename(columns={'B': 'foo', 'C': 'bar'})
.....: )
.....:
Out[142]:
foo bar
A
1 3 0
2 7 3
Deprecate .plotting¶
The pandas.tools.plotting module has been deprecated, in favor of the top level pandas.plotting module. All the public plotting functions are now available
from pandas.plotting (GH12548).
Furthermore, the top-level pandas.scatter_matrix and pandas.plot_params are deprecated.
Users can import these from pandas.plotting as well.
Previous script:
pd.tools.plotting.scatter_matrix(df)
pd.scatter_matrix(df)
Should be changed to:
pd.plotting.scatter_matrix(df)
Other Deprecations¶
SparseArray.to_dense()has deprecated thefillparameter, as that parameter was not being respected (GH14647)SparseSeries.to_dense()has deprecated thesparse_onlyparameter (GH14647)Series.repeat()has deprecated therepsparameter in favor ofrepeats(GH12662)- The
Seriesconstructor and.astypemethod have deprecated accepting timestamp dtypes without a frequency (e.g.np.datetime64) for thedtypeparameter (GH15524) Index.repeat()andMultiIndex.repeat()have deprecated thenparameter in favor ofrepeats(GH12662)Categorical.searchsorted()andSeries.searchsorted()have deprecated thevparameter in favor ofvalue(GH12662)TimedeltaIndex.searchsorted(),DatetimeIndex.searchsorted(), andPeriodIndex.searchsorted()have deprecated thekeyparameter in favor ofvalue(GH12662)DataFrame.astype()has deprecated theraise_on_errorparameter in favor oferrors(GH14878)Series.sortlevelandDataFrame.sortlevelhave been deprecated in favor ofSeries.sort_indexandDataFrame.sort_index(GH15099)- importing
concatfrompandas.tools.mergehas been deprecated in favor of imports from thepandasnamespace. This should only affect explict imports (GH15358) Series/DataFrame/Panel.consolidate()been deprecated as a public method. (GH15483)- The
as_indexerkeyword ofSeries.str.match()has been deprecated (ignored keyword) (GH15257). - The following top-level pandas functions have been deprecated and will be removed in a future version (GH13790, GH15940)
pd.pnow(), replaced byPeriod.now()pd.Term, is removed, as it is not applicable to user code. Instead use in-line string expressions in the where clause when searching in HDFStorepd.Expr, is removed, as it is not applicable to user code.pd.match(), is removed.pd.groupby(), replaced by using the.groupby()method directly on aSeries/DataFramepd.get_store(), replaced by a direct call topd.HDFStore(...)
is_any_int_dtype,is_floating_dtype, andis_sequenceare deprecated frompandas.api.types(GH16042)
Removal of prior version deprecations/changes¶
- The
pandas.rpymodule is removed. Similar functionality can be accessed through the rpy2 project. See the R interfacing docs for more details. - The
pandas.io.gamodule with agoogle-analyticsinterface is removed (GH11308). Similar functionality can be found in the Google2Pandas package. pd.to_datetimeandpd.to_timedeltahave dropped thecoerceparameter in favor oferrors(GH13602)pandas.stats.fama_macbeth,pandas.stats.ols,pandas.stats.plmandpandas.stats.var, as well as the top-levelpandas.fama_macbethandpandas.olsroutines are removed. Similar functionaility can be found in the statsmodels package. (GH11898)- The
TimeSeriesandSparseTimeSeriesclasses, aliases ofSeriesandSparseSeries, are removed (GH10890, GH15098). Series.is_time_seriesis dropped in favor ofSeries.index.is_all_dates(GH15098)- The deprecated
irow,icol,igetandiget_valuemethods are removed in favor ofilocandiatas explained here (GH10711). - The deprecated
DataFrame.iterkv()has been removed in favor ofDataFrame.iteritems()(GH10711) - The
Categoricalconstructor has dropped thenameparameter (GH10632) Categoricalhas dropped support forNaNcategories (GH10748)- The
take_lastparameter has been dropped fromduplicated(),drop_duplicates(),nlargest(), andnsmallest()methods (GH10236, GH10792, GH10920) Series,Index, andDataFramehave dropped thesortandordermethods (GH10726)- Where clauses in
pytablesare only accepted as strings and expressions types and not other data-types (GH12027) DataFramehas dropped thecombineAddandcombineMultmethods in favor ofaddandmulrespectively (GH10735)
Performance Improvements¶
- Improved performance of
pd.wide_to_long()(GH14779) - Improved performance of
pd.factorize()by releasing the GIL withobjectdtype when inferred as strings (GH14859, GH16057) - Improved performance of timeseries plotting with an irregular DatetimeIndex
(or with
compat_x=True) (GH15073). - Improved performance of
groupby().cummin()andgroupby().cummax()(GH15048, GH15109, GH15561, GH15635) - Improved performance and reduced memory when indexing with a
MultiIndex(GH15245) - When reading buffer object in
read_sas()method without specified format, filepath string is inferred rather than buffer object. (GH14947) - Improved performance of
.rank()for categorical data (GH15498) - Improved performance when using
.unstack()(GH15503) - Improved performance of merge/join on
categorycolumns (GH10409) - Improved performance of
drop_duplicates()onboolcolumns (GH12963) - Improve performance of
pd.core.groupby.GroupBy.applywhen the applied function used the.nameattribute of the group DataFrame (GH15062). - Improved performance of
ilocindexing with a list or array (GH15504). - Improved performance of
Series.sort_index()with a monotonic index (GH15694) - Improved performance in
pd.read_csv()on some platforms with buffered reads (GH16039)
Bug Fixes¶
Conversion¶
- Bug in
Timestamp.replacenow raisesTypeErrorwhen incorrect argument names are given; previously this raisedValueError(GH15240) - Bug in
Timestamp.replacewith compat for passing long integers (GH15030) - Bug in
Timestampreturning UTC based time/date attributes when a timezone was provided (GH13303, GH6538) - Bug in
Timestampincorrectly localizing timezones during construction (GH11481, GH15777) - Bug in
TimedeltaIndexaddition where overflow was being allowed without error (GH14816) - Bug in
TimedeltaIndexraising aValueErrorwhen boolean indexing withloc(GH14946) - Bug in catching an overflow in
Timestamp+Timedelta/Offsetoperations (GH15126) - Bug in
DatetimeIndex.round()andTimestamp.round()floating point accuracy when rounding by milliseconds or less (GH14440, GH15578) - Bug in
astype()whereinfvalues were incorrectly converted to integers. Now raises error now withastype()for Series and DataFrames (GH14265) - Bug in
DataFrame(..).apply(to_numeric)when values are of type decimal.Decimal. (GH14827) - Bug in
describe()when passing a numpy array which does not contain the median to thepercentileskeyword argument (GH14908) - Cleaned up
PeriodIndexconstructor, including raising on floats more consistently (GH13277) - Bug in using
__deepcopy__on empty NDFrame objects (GH15370) - Bug in
.replace()may result in incorrect dtypes. (GH12747, GH15765) - Bug in
Series.replaceandDataFrame.replacewhich failed on empty replacement dicts (GH15289) - Bug in
Series.replacewhich replaced a numeric by string (GH15743) - Bug in
Indexconstruction withNaNelements and integer dtype specified (GH15187) - Bug in
Seriesconstruction with a datetimetz (GH14928) - Bug in
Series.dt.round()inconsistent behaviour onNaT‘s with different arguments (GH14940) - Bug in
Seriesconstructor when bothcopy=Trueanddtypearguments are provided (GH15125) - Incorrect dtyped
Serieswas returned by comparison methods (e.g.,lt,gt, ...) against a constant for an emptyDataFrame(GH15077) - Bug in
Series.ffill()with mixed dtypes containing tz-aware datetimes. (GH14956) - Bug in
DataFrame.fillna()where the argumentdowncastwas ignored when fillna value was of typedict(GH15277) - Bug in
.asfreq(), where frequency was not set for emptySeries(GH14320) - Bug in
DataFrameconstruction with nulls and datetimes in a list-like (GH15869) - Bug in
DataFrame.fillna()with tz-aware datetimes (GH15855) - Bug in
is_string_dtype,is_timedelta64_ns_dtype, andis_string_like_dtypein which an error was raised whenNonewas passed in (GH15941) - Bug in the return type of
pd.uniqueon aCategorical, which was returning an ndarray and not aCategorical(GH15903) - Bug in
Index.to_series()where the index was not copied (and so mutating later would change the original), (GH15949) - Bug in indexing with partial string indexing with a len-1 DataFrame (GH16071)
- Bug in
Seriesconstruction where passing invalid dtype didn’t raise an error. (GH15520)
Indexing¶
- Bug in
Indexpower operations with reversed operands (GH14973) - Bug in
DataFrame.sort_values()when sorting by multiple columns where one column is of typeint64and containsNaT(GH14922) - Bug in
DataFrame.reindex()in whichmethodwas ignored when passingcolumns(GH14992) - Bug in
DataFrame.locwith indexing aMultiIndexwith aSeriesindexer (GH14730, GH15424) - Bug in
DataFrame.locwith indexing aMultiIndexwith a numpy array (GH15434) - Bug in
Series.asofwhich raised if the series contained allnp.nan(GH15713) - Bug in
.atwhen selecting from a tz-aware column (GH15822) - Bug in
Series.where()andDataFrame.where()where array-like conditionals were being rejected (GH15414) - Bug in
Series.where()where TZ-aware data was converted to float representation (GH15701) - Bug in
.locthat would not return the correct dtype for scalar access for a DataFrame (GH11617) - Bug in output formatting of a
MultiIndexwhen names are integers (GH12223, GH15262) - Bug in
Categorical.searchsorted()where alphabetical instead of the provided categorical order was used (GH14522) - Bug in
Series.ilocwhere aCategoricalobject for list-like indexes input was returned, where aSerieswas expected. (GH14580) - Bug in
DataFrame.isincomparing datetimelike to empty frame (GH15473) - Bug in
.reset_index()when an allNaNlevel of aMultiIndexwould fail (GH6322) - Bug in
.reset_index()when raising error for index name already present inMultiIndexcolumns (GH16120) - Bug in creating a
MultiIndexwith tuples and not passing a list of names; this will now raiseValueError(GH15110) - Bug in the HTML display with with a
MultiIndexand truncation (GH14882) - Bug in the display of
.info()where a qualifier (+) would always be displayed with aMultiIndexthat contains only non-strings (GH15245) - Bug in
pd.concat()where the names ofMultiIndexof resultingDataFrameare not handled correctly whenNoneis presented in the names ofMultiIndexof inputDataFrame(GH15787) - Bug in
DataFrame.sort_index()andSeries.sort_index()wherena_positiondoesn’t work with aMultiIndex(GH14784, GH16604) - Bug in in
pd.concat()when combining objects with aCategoricalIndex(GH16111) - Bug in indexing with a scalar and a
CategoricalIndex(GH16123)
I/O¶
- Bug in
pd.to_numeric()in which float and unsigned integer elements were being improperly casted (GH14941, GH15005) - Bug in
pd.read_fwf()where the skiprows parameter was not being respected during column width inference (GH11256) - Bug in
pd.read_csv()in which thedialectparameter was not being verified before processing (GH14898) - Bug in
pd.read_csv()in which missing data was being improperly handled withusecols(GH6710) - Bug in
pd.read_csv()in which a file containing a row with many columns followed by rows with fewer columns would cause a crash (GH14125) - Bug in
pd.read_csv()for the C engine whereusecolswere being indexed incorrectly withparse_dates(GH14792) - Bug in
pd.read_csv()withparse_dateswhen multiline headers are specified (GH15376) - Bug in
pd.read_csv()withfloat_precision='round_trip'which caused a segfault when a text entry is parsed (GH15140) - Bug in
pd.read_csv()when an index was specified and no values were specified as null values (GH15835) - Bug in
pd.read_csv()in which certain invalid file objects caused the Python interpreter to crash (GH15337) - Bug in
pd.read_csv()in which invalid values fornrowsandchunksizewere allowed (GH15767) - Bug in
pd.read_csv()for the Python engine in which unhelpful error messages were being raised when parsing errors occurred (GH15910) - Bug in
pd.read_csv()in which theskipfooterparameter was not being properly validated (GH15925) - Bug in
pd.to_csv()in which there was numeric overflow when a timestamp index was being written (GH15982) - Bug in
pd.util.hashing.hash_pandas_object()in which hashing of categoricals depended on the ordering of categories, instead of just their values. (GH15143) - Bug in
.to_json()wherelines=Trueand contents (keys or values) contain escaped characters (GH15096) - Bug in
.to_json()causing single byte ascii characters to be expanded to four byte unicode (GH15344) - Bug in
.to_json()for the C engine where rollover was not correctly handled for case where frac is odd and diff is exactly 0.5 (GH15716, GH15864) - Bug in
pd.read_json()for Python 2 wherelines=Trueand contents contain non-ascii unicode characters (GH15132) - Bug in
pd.read_msgpack()in whichSeriescategoricals were being improperly processed (GH14901) - Bug in
pd.read_msgpack()which did not allow loading of a dataframe with an index of typeCategoricalIndex(GH15487) - Bug in
pd.read_msgpack()when deserializing aCategoricalIndex(GH15487) - Bug in
DataFrame.to_records()with converting aDatetimeIndexwith a timezone (GH13937) - Bug in
DataFrame.to_records()which failed with unicode characters in column names (GH11879) - Bug in
.to_sql()when writing a DataFrame with numeric index names (GH15404). - Bug in
DataFrame.to_html()withindex=Falseandmax_rowsraising inIndexError(GH14998) - Bug in
pd.read_hdf()passing aTimestampto thewhereparameter with a non date column (GH15492) - Bug in
DataFrame.to_stata()andStataWriterwhich produces incorrectly formatted files to be produced for some locales (GH13856) - Bug in
StataReaderandStataWriterwhich allows invalid encodings (GH15723) - Bug in the
Seriesrepr not showing the length when the output was truncated (GH15962).
Plotting¶
- Bug in
DataFrame.histwhereplt.tight_layoutcaused anAttributeError(usematplotlib >= 2.0.1) (GH9351) - Bug in
DataFrame.boxplotwherefontsizewas not applied to the tick labels on both axes (GH15108) - Bug in the date and time converters pandas registers with matplotlib not handling multiple dimensions (GH16026)
- Bug in
pd.scatter_matrix()could accept eithercolororc, but not both (GH14855)
Groupby/Resample/Rolling¶
- Bug in
.groupby(..).resample()when passed theon=kwarg. (GH15021) - Properly set
__name__and__qualname__forGroupby.*functions (GH14620) - Bug in
GroupBy.get_group()failing with a categorical grouper (GH15155) - Bug in
.groupby(...).rolling(...)whenonis specified and using aDatetimeIndex(GH15130, GH13966) - Bug in groupby operations with
timedelta64when passingnumeric_only=False(GH5724) - Bug in
groupby.apply()coercingobjectdtypes to numeric types, when not all values were numeric (GH14423, GH15421, GH15670) - Bug in
resample, where a non-stringloffsetargument would not be applied when resampling a timeseries (GH13218) - Bug in
DataFrame.groupby().describe()when grouping onIndexcontaining tuples (GH14848) - Bug in
groupby().nunique()with a datetimelike-grouper where bins counts were incorrect (GH13453) - Bug in
groupby.transform()that would coerce the resultant dtypes back to the original (GH10972, GH11444) - Bug in
groupby.agg()incorrectly localizing timezone ondatetime(GH15426, GH10668, GH13046) - Bug in
.rolling/expanding()functions wherecount()was not countingnp.Inf, nor handlingobjectdtypes (GH12541) - Bug in
.rolling()wherepd.Timedeltaordatetime.timedeltawas not accepted as awindowargument (GH15440) - Bug in
Rolling.quantilefunction that caused a segmentation fault when called with a quantile value outside of the range [0, 1] (GH15463) - Bug in
DataFrame.resample().median()if duplicate column names are present (GH14233)
Sparse¶
- Bug in
SparseSeries.reindexon single level with list of length 1 (GH15447) - Bug in repr-formatting a
SparseDataFrameafter a value was set on (a copy of) one of its series (GH15488) - Bug in
SparseDataFrameconstruction with lists not coercing to dtype (GH15682) - Bug in sparse array indexing in which indices were not being validated (GH15863)
Reshaping¶
- Bug in
pd.merge_asof()whereleft_indexorright_indexcaused a failure when multiplebywas specified (GH15676) - Bug in
pd.merge_asof()whereleft_index/right_indextogether caused a failure whentolerancewas specified (GH15135) - Bug in
DataFrame.pivot_table()wheredropna=Truewould not drop all-NaN columns when the columns was acategorydtype (GH15193) - Bug in
pd.melt()where passing a tuple value forvalue_varscaused aTypeError(GH15348) - Bug in
pd.pivot_table()where no error was raised when values argument was not in the columns (GH14938) - Bug in
pd.concat()in which concatenating with an empty dataframe withjoin='inner'was being improperly handled (GH15328) - Bug with
sort=TrueinDataFrame.joinandpd.mergewhen joining on indexes (GH15582) - Bug in
DataFrame.nsmallestandDataFrame.nlargestwhere identical values resulted in duplicated rows (GH15297)
Numeric¶
- Bug in
.rank()which incorrectly ranks ordered categories (GH15420) - Bug in
.corr()and.cov()where the column and index were the same object (GH14617) - Bug in
.mode()wheremodewas not returned if was only a single value (GH15714) - Bug in
pd.cut()with a single bin on an all 0s array (GH15428) - Bug in
pd.qcut()with a single quantile and an array with identical values (GH15431) - Bug in
pandas.tools.utils.cartesian_product()with large input can cause overflow on windows (GH15265) - Bug in
.eval()which caused multiline evals to fail with local variables not on the first line (GH15342)
Other¶
- Compat with SciPy 0.19.0 for testing on
.interpolate()(GH15662) - Compat for 32-bit platforms for
.qcut/cut; bins will now beint64dtype (GH14866) - Bug in interactions with
Qtwhen aQtApplicationalready exists (GH14372) - Avoid use of
np.finfo()duringimport pandasremoved to mitigate deadlock on Python GIL misuse (GH14641)
v0.19.2 (December 24, 2016)¶
This is a minor bug-fix release in the 0.19.x series and includes some small regression fixes, bug fixes and performance improvements. We recommend that all users upgrade to this version.
Highlights include:
- Compatibility with Python 3.6
- Added a Pandas Cheat Sheet. (GH13202).
What’s new in v0.19.2
Enhancements¶
The pd.merge_asof(), added in 0.19.0, gained some improvements:
Performance Improvements¶
Bug Fixes¶
- Compat with python 3.6 for pickling of some offsets (GH14685)
- Compat with python 3.6 for some indexing exception types (GH14684, GH14689)
- Compat with python 3.6 for deprecation warnings in the test suite (GH14681)
- Compat with python 3.6 for Timestamp pickles (GH14689)
- Compat with
dateutil==2.6.0; segfault reported in the testing suite (GH14621) - Allow
nanosecondsinTimestamp.replaceas a kwarg (GH14621) - Bug in
pd.read_csvin which aliasing was being done forna_valueswhen passed in as a dictionary (GH14203) - Bug in
pd.read_csvin which column indices for a dict-likena_valueswere not being respected (GH14203) - Bug in
pd.read_csvwhere reading files fails, if the number of headers is equal to the number of lines in the file (GH14515) - Bug in
pd.read_csvfor the Python engine in which an unhelpful error message was being raised when multi-char delimiters were not being respected with quotes (GH14582) - Fix bugs (GH14734, GH13654) in
pd.read_sasandpandas.io.sas.sas7bdat.SAS7BDATReaderthat caused problems when reading a SAS file incrementally. - Bug in
pd.read_csvfor the Python engine in which an unhelpful error message was being raised whenskipfooterwas not being respected by Python’s CSV library (GH13879) - Bug in
.fillna()in which timezone aware datetime64 values were incorrectly rounded (GH14872) - Bug in
.groupby(..., sort=True)of a non-lexsorted MultiIndex when grouping with multiple levels (GH14776) - Bug in
pd.cutwith negative values and a single bin (GH14652) - Bug in
pd.to_numericwhere a 0 was not unsigned on adowncast='unsigned'argument (GH14401) - Bug in plotting regular and irregular timeseries using shared axes
(
sharex=Trueorax.twinx()) (GH13341, GH14322). - Bug in not propogating exceptions in parsing invalid datetimes, noted in python 3.6 (GH14561)
- Bug in resampling a
DatetimeIndexin local TZ, covering a DST change, which would raiseAmbiguousTimeError(GH14682) - Bug in indexing that transformed
RecursionErrorintoKeyErrororIndexingError(GH14554) - Bug in
HDFStorewhen writing aMultiIndexwhen usingdata_columns=True(GH14435) - Bug in
HDFStore.append()when writing aSeriesand passing amin_itemsizeargument containing a value for theindex(GH11412) - Bug when writing to a
HDFStoreintableformat with amin_itemsizevalue for theindexand without asking to append (GH10381) - Bug in
Series.groupby.nunique()raising anIndexErrorfor an emptySeries(GH12553) - Bug in
DataFrame.nlargestandDataFrame.nsmallestwhen the index had duplicate values (GH13412) - Bug in clipboard functions on linux with python2 with unicode and separators (GH13747)
- Bug in clipboard functions on Windows 10 and python 3 (GH14362, GH12807)
- Bug in
.to_clipboard()and Excel compat (GH12529) - Bug in
DataFrame.combine_first()for integer columns (GH14687). - Bug in
pd.read_csv()in which thedtypeparameter was not being respected for empty data (GH14712) - Bug in
pd.read_csv()in which thenrowsparameter was not being respected for large input when using the C engine for parsing (GH7626) - Bug in
pd.merge_asof()could not handle timezone-aware DatetimeIndex when a tolerance was specified (GH14844) - Explicit check in
to_stataandStataWriterfor out-of-range values when writing doubles (GH14618) - Bug in
.plot(kind='kde')which did not drop missing values to generate the KDE Plot, instead generating an empty plot. (GH14821) - Bug in
unstack()if called with a list of column(s) as an argument, regardless of the dtypes of all columns, they get coerced toobject(GH11847)
v0.19.1 (November 3, 2016)¶
This is a minor bug-fix release from 0.19.0 and includes some small regression fixes, bug fixes and performance improvements. We recommend that all users upgrade to this version.
What’s new in v0.19.1
Performance Improvements¶
- Fixed performance regression in factorization of
Perioddata (GH14338) - Fixed performance regression in
Series.asof(where)whenwhereis a scalar (GH14461) - Improved performance in
DataFrame.asof(where)whenwhereis a scalar (GH14461) - Improved performance in
.to_json()whenlines=True(GH14408) - Improved performance in certain types of loc indexing with a MultiIndex (GH14551).
Bug Fixes¶
- Source installs from PyPI will now again work without
cythoninstalled, as in previous versions (GH14204) - Compat with Cython 0.25 for building (GH14496)
- Fixed regression where user-provided file handles were closed in
read_csv(c engine) (GH14418). - Fixed regression in
DataFrame.quantilewhen missing values where present in some columns (GH14357). - Fixed regression in
Index.differencewhere thefreqof aDatetimeIndexwas incorrectly set (GH14323) - Added back
pandas.core.common.array_equivalentwith a deprecation warning (GH14555). - Bug in
pd.read_csvfor the C engine in which quotation marks were improperly parsed in skipped rows (GH14459) - Bug in
pd.read_csvfor Python 2.x in which Unicode quote characters were no longer being respected (GH14477) - Fixed regression in
Index.appendwhen categorical indices were appended (GH14545). - Fixed regression in
pd.DataFramewhere constructor fails when given dict withNonevalue (GH14381) - Fixed regression in
DatetimeIndex._maybe_cast_slice_boundwhen index is empty (GH14354). - Bug in localizing an ambiguous timezone when a boolean is passed (GH14402)
- Bug in
TimedeltaIndexaddition with a Datetime-like object where addition overflow in the negative direction was not being caught (GH14068, GH14453) - Bug in string indexing against data with
objectIndexmay raiseAttributeError(GH14424) - Corrrecly raise
ValueErroron empty input topd.eval()anddf.query()(GH13139) - Bug in
RangeIndex.intersectionwhen result is a empty set (GH14364). - Bug in groupby-transform broadcasting that could cause incorrect dtype coercion (GH14457)
- Bug in
Series.__setitem__which allowed mutating read-only arrays (GH14359). - Bug in
DataFrame.insertwhere multiple calls with duplicate columns can fail (GH14291) pd.merge()will raiseValueErrorwith non-boolean parameters in passed boolean type arguments (GH14434)- Bug in
Timestampwhere dates very near the minimum (1677-09) could underflow on creation (GH14415) - Bug in
pd.concatwhere names of thekeyswere not propagated to the resultingMultiIndex(GH14252) - Bug in
pd.concatwhereaxiscannot take string parameters'rows'or'columns'(GH14369) - Bug in
pd.concatwith dataframes heterogeneous in length and tuplekeys(GH14438) - Bug in
MultiIndex.set_levelswhere illegal level values were still set after raising an error (GH13754) - Bug in
DataFrame.to_jsonwherelines=Trueand a value contained a}character (GH14391) - Bug in
df.groupbycausing anAttributeErrorwhen grouping a single index frame by a column and the index level (:issue`14327`) - Bug in
df.groupbywhereTypeErrorraised whenpd.Grouper(key=...)is passed in a list (GH14334) - Bug in
pd.pivot_tablemay raiseTypeErrororValueErrorwhenindexorcolumnsis not scalar andvaluesis not specified (GH14380)
v0.19.0 (October 2, 2016)¶
This is a major release from 0.18.1 and includes number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
Highlights include:
merge_asof()for asof-style time-series joining, see here.rolling()is now time-series aware, see hereread_csv()now supports parsingCategoricaldata, see here- A function
union_categorical()has been added for combining categoricals, see here PeriodIndexnow has its ownperioddtype, and changed to be more consistent with otherIndexclasses. See here- Sparse data structures gained enhanced support of
intandbooldtypes, see here - Comparison operations with
Seriesno longer ignores the index, see here for an overview of the API changes. - Introduction of a pandas development API for utility functions, see here.
- Deprecation of
Panel4DandPanelND. We recommend to represent these types of n-dimensional data with the xarray package. - Removal of the previously deprecated modules
pandas.io.data,pandas.io.wb,pandas.tools.rplot.
Warning
pandas >= 0.19.0 will no longer silence numpy ufunc warnings upon import, see here.
What’s new in v0.19.0
- New features
merge_asoffor asof-style time-series joining.rolling()is now time-series awareread_csvhas improved support for duplicate column namesread_csvsupports parsingCategoricaldirectly- Categorical Concatenation
- Semi-Month Offsets
- New Index methods
- Google BigQuery Enhancements
- Fine-grained numpy errstate
get_dummiesnow returns integer dtypes- Downcast values to smallest possible dtype in
to_numeric - pandas development API
- Other enhancements
- API changes
Series.tolist()will now return Python typesSeriesoperators for different indexesSeriestype promotion on assignment.to_datetime()changes- Merging changes
.describe()changesPeriodchanges- Index
+/-no longer used for set operations Index.differenceand.symmetric_differencechangesIndex.uniqueconsistently returnsIndexMultiIndexconstructors,groupbyandset_indexpreserve categorical dtypesread_csvwill progressively enumerate chunks- Sparse Changes
- Indexer dtype changes
- Other API Changes
- Deprecations
- Removal of prior version deprecations/changes
- Performance Improvements
- Bug Fixes
New features¶
merge_asof for asof-style time-series joining¶
A long-time requested feature has been added through the merge_asof() function, to
support asof style joining of time-series (GH1870, GH13695, GH13709, GH13902). Full documentation is
here.
The merge_asof() performs an asof merge, which is similar to a left-join
except that we match on nearest key rather than equal keys.
In [1]: left = pd.DataFrame({'a': [1, 5, 10],
...: 'left_val': ['a', 'b', 'c']})
...:
In [2]: right = pd.DataFrame({'a': [1, 2, 3, 6, 7],
...: 'right_val': [1, 2, 3, 6, 7]})
...:
In [3]: left
Out[3]:
a left_val
0 1 a
1 5 b
2 10 c
In [4]: right
���������������������������������������������������������������������Out[4]:
a right_val
0 1 1
1 2 2
2 3 3
3 6 6
4 7 7
We typically want to match exactly when possible, and use the most recent value otherwise.
In [5]: pd.merge_asof(left, right, on='a')
Out[5]:
a left_val right_val
0 1 a 1
1 5 b 3
2 10 c 7
We can also match rows ONLY with prior data, and not an exact match.
In [6]: pd.merge_asof(left, right, on='a', allow_exact_matches=False)
Out[6]:
a left_val right_val
0 1 a NaN
1 5 b 3.0
2 10 c 7.0
In a typical time-series example, we have trades and quotes and we want to asof-join them.
This also illustrates using the by parameter to group data before merging.
In [7]: trades = pd.DataFrame({
...: 'time': pd.to_datetime(['20160525 13:30:00.023',
...: '20160525 13:30:00.038',
...: '20160525 13:30:00.048',
...: '20160525 13:30:00.048',
...: '20160525 13:30:00.048']),
...: 'ticker': ['MSFT', 'MSFT',
...: 'GOOG', 'GOOG', 'AAPL'],
...: 'price': [51.95, 51.95,
...: 720.77, 720.92, 98.00],
...: 'quantity': [75, 155,
...: 100, 100, 100]},
...: columns=['time', 'ticker', 'price', 'quantity'])
...:
In [8]: quotes = pd.DataFrame({
...: 'time': pd.to_datetime(['20160525 13:30:00.023',
...: '20160525 13:30:00.023',
...: '20160525 13:30:00.030',
...: '20160525 13:30:00.041',
...: '20160525 13:30:00.048',
...: '20160525 13:30:00.049',
...: '20160525 13:30:00.072',
...: '20160525 13:30:00.075']),
...: 'ticker': ['GOOG', 'MSFT', 'MSFT',
...: 'MSFT', 'GOOG', 'AAPL', 'GOOG',
...: 'MSFT'],
...: 'bid': [720.50, 51.95, 51.97, 51.99,
...: 720.50, 97.99, 720.50, 52.01],
...: 'ask': [720.93, 51.96, 51.98, 52.00,
...: 720.93, 98.01, 720.88, 52.03]},
...: columns=['time', 'ticker', 'bid', 'ask'])
...:
In [9]: trades
Out[9]:
time ticker price quantity
0 2016-05-25 13:30:00.023 MSFT 51.95 75
1 2016-05-25 13:30:00.038 MSFT 51.95 155
2 2016-05-25 13:30:00.048 GOOG 720.77 100
3 2016-05-25 13:30:00.048 GOOG 720.92 100
4 2016-05-25 13:30:00.048 AAPL 98.00 100
In [10]: quotes
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[10]:
time ticker bid ask
0 2016-05-25 13:30:00.023 GOOG 720.50 720.93
1 2016-05-25 13:30:00.023 MSFT 51.95 51.96
2 2016-05-25 13:30:00.030 MSFT 51.97 51.98
3 2016-05-25 13:30:00.041 MSFT 51.99 52.00
4 2016-05-25 13:30:00.048 GOOG 720.50 720.93
5 2016-05-25 13:30:00.049 AAPL 97.99 98.01
6 2016-05-25 13:30:00.072 GOOG 720.50 720.88
7 2016-05-25 13:30:00.075 MSFT 52.01 52.03
An asof merge joins on the on, typically a datetimelike field, which is ordered, and
in this case we are using a grouper in the by field. This is like a left-outer join, except
that forward filling happens automatically taking the most recent non-NaN value.
In [11]: pd.merge_asof(trades, quotes,
....: on='time',
....: by='ticker')
....:
Out[11]:
time ticker price quantity bid ask
0 2016-05-25 13:30:00.023 MSFT 51.95 75 51.95 51.96
1 2016-05-25 13:30:00.038 MSFT 51.95 155 51.97 51.98
2 2016-05-25 13:30:00.048 GOOG 720.77 100 720.50 720.93
3 2016-05-25 13:30:00.048 GOOG 720.92 100 720.50 720.93
4 2016-05-25 13:30:00.048 AAPL 98.00 100 NaN NaN
This returns a merged DataFrame with the entries in the same order as the original left
passed DataFrame (trades in this case), with the fields of the quotes merged.
.rolling() is now time-series aware¶
.rolling() objects are now time-series aware and can accept a time-series offset (or convertible) for the window argument (GH13327, GH12995).
See the full documentation here.
In [12]: dft = pd.DataFrame({'B': [0, 1, 2, np.nan, 4]},
....: index=pd.date_range('20130101 09:00:00', periods=5, freq='s'))
....:
In [13]: dft
Out[13]:
B
2013-01-01 09:00:00 0.0
2013-01-01 09:00:01 1.0
2013-01-01 09:00:02 2.0
2013-01-01 09:00:03 NaN
2013-01-01 09:00:04 4.0
This is a regular frequency index. Using an integer window parameter works to roll along the window frequency.
In [14]: dft.rolling(2).sum()
Out[14]:
B
2013-01-01 09:00:00 NaN
2013-01-01 09:00:01 1.0
2013-01-01 09:00:02 3.0
2013-01-01 09:00:03 NaN
2013-01-01 09:00:04 NaN
In [15]: dft.rolling(2, min_periods=1).sum()
����������������������������������������������������������������������������������������������������������������������������������������������������������������Out[15]:
B
2013-01-01 09:00:00 0.0
2013-01-01 09:00:01 1.0
2013-01-01 09:00:02 3.0
2013-01-01 09:00:03 2.0
2013-01-01 09:00:04 4.0
Specifying an offset allows a more intuitive specification of the rolling frequency.
In [16]: dft.rolling('2s').sum()
Out[16]:
B
2013-01-01 09:00:00 0.0
2013-01-01 09:00:01 1.0
2013-01-01 09:00:02 3.0
2013-01-01 09:00:03 2.0
2013-01-01 09:00:04 4.0
Using a non-regular, but still monotonic index, rolling with an integer window does not impart any special calculation.
In [17]: dft = DataFrame({'B': [0, 1, 2, np.nan, 4]},
....: index = pd.Index([pd.Timestamp('20130101 09:00:00'),
....: pd.Timestamp('20130101 09:00:02'),
....: pd.Timestamp('20130101 09:00:03'),
....: pd.Timestamp('20130101 09:00:05'),
....: pd.Timestamp('20130101 09:00:06')],
....: name='foo'))
....:
In [18]: dft
Out[18]:
B
foo
2013-01-01 09:00:00 0.0
2013-01-01 09:00:02 1.0
2013-01-01 09:00:03 2.0
2013-01-01 09:00:05 NaN
2013-01-01 09:00:06 4.0
In [19]: dft.rolling(2).sum()
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[19]:
B
foo
2013-01-01 09:00:00 NaN
2013-01-01 09:00:02 1.0
2013-01-01 09:00:03 3.0
2013-01-01 09:00:05 NaN
2013-01-01 09:00:06 NaN
Using the time-specification generates variable windows for this sparse data.
In [20]: dft.rolling('2s').sum()
Out[20]:
B
foo
2013-01-01 09:00:00 0.0
2013-01-01 09:00:02 1.0
2013-01-01 09:00:03 3.0
2013-01-01 09:00:05 NaN
2013-01-01 09:00:06 4.0
Furthermore, we now allow an optional on parameter to specify a column (rather than the
default of the index) in a DataFrame.
In [21]: dft = dft.reset_index()
In [22]: dft
Out[22]:
foo B
0 2013-01-01 09:00:00 0.0
1 2013-01-01 09:00:02 1.0
2 2013-01-01 09:00:03 2.0
3 2013-01-01 09:00:05 NaN
4 2013-01-01 09:00:06 4.0
In [23]: dft.rolling('2s', on='foo').sum()
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[23]:
foo B
0 2013-01-01 09:00:00 0.0
1 2013-01-01 09:00:02 1.0
2 2013-01-01 09:00:03 3.0
3 2013-01-01 09:00:05 NaN
4 2013-01-01 09:00:06 4.0
read_csv has improved support for duplicate column names¶
Duplicate column names are now supported in read_csv() whether
they are in the file or passed in as the names parameter (GH7160, GH9424)
In [24]: data = '0,1,2\n3,4,5'
In [25]: names = ['a', 'b', 'a']
Previous behavior:
In [2]: pd.read_csv(StringIO(data), names=names)
Out[2]:
a b a
0 2 1 2
1 5 4 5
The first a column contained the same data as the second a column, when it should have
contained the values [0, 3].
New behavior:
In [26]: pd.read_csv(StringIO(data), names=names)
Out[26]:
a b a.1
0 0 1 2
1 3 4 5
read_csv supports parsing Categorical directly¶
The read_csv() function now supports parsing a Categorical column when
specified as a dtype (GH10153). Depending on the structure of the data,
this can result in a faster parse time and lower memory usage compared to
converting to Categorical after parsing. See the io docs here.
In [27]: data = 'col1,col2,col3\na,b,1\na,b,2\nc,d,3'
In [28]: pd.read_csv(StringIO(data))
Out[28]:
col1 col2 col3
0 a b 1
1 a b 2
2 c d 3
In [29]: pd.read_csv(StringIO(data)).dtypes
����������������������������������������������������������������������������������Out[29]:
col1 object
col2 object
col3 int64
dtype: object
In [30]: pd.read_csv(StringIO(data), dtype='category').dtypes
�������������������������������������������������������������������������������������������������������������������������������������������������������Out[30]:
col1 category
col2 category
col3 category
dtype: object
Individual columns can be parsed as a Categorical using a dict specification
In [31]: pd.read_csv(StringIO(data), dtype={'col1': 'category'}).dtypes
Out[31]:
col1 category
col2 object
col3 int64
dtype: object
Note
The resulting categories will always be parsed as strings (object dtype).
If the categories are numeric they can be converted using the
to_numeric() function, or as appropriate, another converter
such as to_datetime().
In [32]: df = pd.read_csv(StringIO(data), dtype='category')
In [33]: df.dtypes
Out[33]:
col1 category
col2 category
col3 category
dtype: object
In [34]: df['col3']
���������������������������������������������������������������������������Out[34]:
0 1
1 2
2 3
Name: col3, dtype: category
Categories (3, object): [1, 2, 3]
In [35]: df['col3'].cat.categories = pd.to_numeric(df['col3'].cat.categories)
In [36]: df['col3']
Out[36]:
0 1
1 2
2 3
Name: col3, dtype: category
Categories (3, int64): [1, 2, 3]
Categorical Concatenation¶
A function
union_categoricals()has been added for combining categoricals, see Unioning Categoricals (GH13361, GH:13763, issue:13846, GH14173)In [37]: from pandas.api.types import union_categoricals In [38]: a = pd.Categorical(["b", "c"]) In [39]: b = pd.Categorical(["a", "b"]) In [40]: union_categoricals([a, b]) Out[40]: [b, c, a, b] Categories (3, object): [b, c, a]
concatandappendnow can concatcategorydtypes with differentcategoriesasobjectdtype (GH13524)In [41]: s1 = pd.Series(['a', 'b'], dtype='category') In [42]: s2 = pd.Series(['b', 'c'], dtype='category')
Previous behavior:
In [1]: pd.concat([s1, s2]) ValueError: incompatible categories in categorical concat
New behavior:
In [43]: pd.concat([s1, s2]) Out[43]: 0 a 1 b 0 b 1 c dtype: object
Semi-Month Offsets¶
Pandas has gained new frequency offsets, SemiMonthEnd (‘SM’) and SemiMonthBegin (‘SMS’).
These provide date offsets anchored (by default) to the 15th and end of month, and 15th and 1st of month respectively.
(GH1543)
In [44]: from pandas.tseries.offsets import SemiMonthEnd, SemiMonthBegin
SemiMonthEnd:
In [45]: Timestamp('2016-01-01') + SemiMonthEnd()
Out[45]: Timestamp('2016-01-15 00:00:00')
In [46]: pd.date_range('2015-01-01', freq='SM', periods=4)
������������������������������������������Out[46]: DatetimeIndex(['2015-01-15', '2015-01-31', '2015-02-15', '2015-02-28'], dtype='datetime64[ns]', freq='SM-15')
SemiMonthBegin:
In [47]: Timestamp('2016-01-01') + SemiMonthBegin()
Out[47]: Timestamp('2016-01-15 00:00:00')
In [48]: pd.date_range('2015-01-01', freq='SMS', periods=4)
������������������������������������������Out[48]: DatetimeIndex(['2015-01-01', '2015-01-15', '2015-02-01', '2015-02-15'], dtype='datetime64[ns]', freq='SMS-15')
Using the anchoring suffix, you can also specify the day of month to use instead of the 15th.
In [49]: pd.date_range('2015-01-01', freq='SMS-16', periods=4)
Out[49]: DatetimeIndex(['2015-01-01', '2015-01-16', '2015-02-01', '2015-02-16'], dtype='datetime64[ns]', freq='SMS-16')
In [50]: pd.date_range('2015-01-01', freq='SM-14', periods=4)
������������������������������������������������������������������������������������������������������������������������Out[50]: DatetimeIndex(['2015-01-14', '2015-01-31', '2015-02-14', '2015-02-28'], dtype='datetime64[ns]', freq='SM-14')
New Index methods¶
The following methods and options are added to Index, to be more consistent with the Series and DataFrame API.
Index now supports the .where() function for same shape indexing (GH13170)
In [51]: idx = pd.Index(['a', 'b', 'c'])
In [52]: idx.where([True, False, True])
Out[52]: Index(['a', nan, 'c'], dtype='object')
Index now supports .dropna() to exclude missing values (GH6194)
In [53]: idx = pd.Index([1, 2, np.nan, 4])
In [54]: idx.dropna()
Out[54]: Float64Index([1.0, 2.0, 4.0], dtype='float64')
For MultiIndex, values are dropped if any level is missing by default. Specifying
how='all' only drops values where all levels are missing.
In [55]: midx = pd.MultiIndex.from_arrays([[1, 2, np.nan, 4],
....: [1, 2, np.nan, np.nan]])
....:
In [56]: midx
Out[56]:
MultiIndex(levels=[[1, 2, 4], [1, 2]],
labels=[[0, 1, -1, 2], [0, 1, -1, -1]])
In [57]: midx.dropna()
����������������������������������������������������������������������������������������������������Out[57]:
MultiIndex(levels=[[1, 2, 4], [1, 2]],
labels=[[0, 1], [0, 1]])
In [58]: midx.dropna(how='all')
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[58]:
MultiIndex(levels=[[1, 2, 4], [1, 2]],
labels=[[0, 1, 2], [0, 1, -1]])
Index now supports .str.extractall() which returns a DataFrame, see the docs here (GH10008, GH13156)
In [59]: idx = pd.Index(["a1a2", "b1", "c1"])
In [60]: idx.str.extractall("[ab](?P<digit>\d)")
Out[60]:
digit
match
0 0 1
1 2
1 0 1
Index.astype() now accepts an optional boolean argument copy, which allows optional copying if the requirements on dtype are satisfied (GH13209)
Google BigQuery Enhancements¶
- The
read_gbq()method has gained thedialectargument to allow users to specify whether to use BigQuery’s legacy SQL or BigQuery’s standard SQL. See the docs for more details (GH13615). - The
to_gbq()method now allows the DataFrame column order to differ from the destination table schema (GH11359).
Fine-grained numpy errstate¶
Previous versions of pandas would permanently silence numpy’s ufunc error handling when pandas was imported. Pandas did this in order to silence the warnings that would arise from using numpy ufuncs on missing data, which are usually represented as NaN s. Unfortunately, this silenced legitimate warnings arising in non-pandas code in the application. Starting with 0.19.0, pandas will use the numpy.errstate context manager to silence these warnings in a more fine-grained manner, only around where these operations are actually used in the pandas codebase. (GH13109, GH13145)
After upgrading pandas, you may see new RuntimeWarnings being issued from your code. These are likely legitimate, and the underlying cause likely existed in the code when using previous versions of pandas that simply silenced the warning. Use numpy.errstate around the source of the RuntimeWarning to control how these conditions are handled.
get_dummies now returns integer dtypes¶
The pd.get_dummies function now returns dummy-encoded columns as small integers, rather than floats (GH8725). This should provide an improved memory footprint.
Previous behavior:
In [1]: pd.get_dummies(['a', 'b', 'a', 'c']).dtypes
Out[1]:
a float64
b float64
c float64
dtype: object
New behavior:
In [61]: pd.get_dummies(['a', 'b', 'a', 'c']).dtypes
Out[61]:
a uint8
b uint8
c uint8
dtype: object
Downcast values to smallest possible dtype in to_numeric¶
pd.to_numeric() now accepts a downcast parameter, which will downcast the data if possible to smallest specified numerical dtype (GH13352)
In [62]: s = ['1', 2, 3]
In [63]: pd.to_numeric(s, downcast='unsigned')
Out[63]: array([1, 2, 3], dtype=uint8)
In [64]: pd.to_numeric(s, downcast='integer')
���������������������������������������Out[64]: array([1, 2, 3], dtype=int8)
pandas development API¶
As part of making pandas API more uniform and accessible in the future, we have created a standard
sub-package of pandas, pandas.api to hold public API’s. We are starting by exposing type
introspection functions in pandas.api.types. More sub-packages and officially sanctioned API’s
will be published in future versions of pandas (GH13147, GH13634)
The following are now part of this API:
In [65]: import pprint
In [66]: from pandas.api import types
In [67]: funcs = [ f for f in dir(types) if not f.startswith('_') ]
In [68]: pprint.pprint(funcs)
['CategoricalDtype',
'DatetimeTZDtype',
'IntervalDtype',
'PeriodDtype',
'infer_dtype',
'is_any_int_dtype',
'is_bool',
'is_bool_dtype',
'is_categorical',
'is_categorical_dtype',
'is_complex',
'is_complex_dtype',
'is_datetime64_any_dtype',
'is_datetime64_dtype',
'is_datetime64_ns_dtype',
'is_datetime64tz_dtype',
'is_datetimetz',
'is_dict_like',
'is_dtype_equal',
'is_extension_type',
'is_file_like',
'is_float',
'is_float_dtype',
'is_floating_dtype',
'is_hashable',
'is_int64_dtype',
'is_integer',
'is_integer_dtype',
'is_interval',
'is_interval_dtype',
'is_iterator',
'is_list_like',
'is_named_tuple',
'is_number',
'is_numeric_dtype',
'is_object_dtype',
'is_period',
'is_period_dtype',
'is_re',
'is_re_compilable',
'is_scalar',
'is_sequence',
'is_signed_integer_dtype',
'is_sparse',
'is_string_dtype',
'is_timedelta64_dtype',
'is_timedelta64_ns_dtype',
'is_unsigned_integer_dtype',
'pandas_dtype',
'union_categoricals']
Note
Calling these functions from the internal module pandas.core.common will now show a DeprecationWarning (GH13990)
Other enhancements¶
Timestampcan now accept positional and keyword parameters similar todatetime.datetime()(GH10758, GH11630)In [69]: pd.Timestamp(2012, 1, 1) Out[69]: Timestamp('2012-01-01 00:00:00') In [70]: pd.Timestamp(year=2012, month=1, day=1, hour=8, minute=30) ������������������������������������������Out[70]: Timestamp('2012-01-01 08:30:00')
The
.resample()function now accepts aon=orlevel=parameter for resampling on a datetimelike column orMultiIndexlevel (GH13500)In [71]: df = pd.DataFrame({'date': pd.date_range('2015-01-01', freq='W', periods=5), ....: 'a': np.arange(5)}, ....: index=pd.MultiIndex.from_arrays([ ....: [1,2,3,4,5], ....: pd.date_range('2015-01-01', freq='W', periods=5)], ....: names=['v','d'])) ....: In [72]: df Out[72]: a date v d 1 2015-01-04 0 2015-01-04 2 2015-01-11 1 2015-01-11 3 2015-01-18 2 2015-01-18 4 2015-01-25 3 2015-01-25 5 2015-02-01 4 2015-02-01 In [73]: df.resample('M', on='date').sum() �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[73]: a date 2015-01-31 6 2015-02-28 4 In [74]: df.resample('M', level='d').sum() �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[74]: a d 2015-01-31 6 2015-02-28 4
The
.get_credentials()method ofGbqConnectorcan now first try to fetch the application default credentials. See the docs for more details (GH13577).The
.tz_localize()method ofDatetimeIndexandTimestamphas gained theerrorskeyword, so you can potentially coerce nonexistent timestamps toNaT. The default behavior remains to raising aNonExistentTimeError(GH13057).to_hdf/read_hdf()now accept path objects (e.g.pathlib.Path,py.path.local) for the file path (GH11773)The
pd.read_csv()withengine='python'has gained support for thedecimal(GH12933),na_filter(GH13321) and thememory_mapoption (GH13381).Consistent with the Python API,
pd.read_csv()will now interpret+infas positive infinity (GH13274)The
pd.read_html()has gained support for thena_values,converters,keep_default_naoptions (GH13461)Categorical.astype()now accepts an optional boolean argumentcopy, effective when dtype is categorical (GH13209)DataFramehas gained the.asof()method to return the last non-NaN values according to the selected subset (GH13358)The
DataFrameconstructor will now respect key ordering if a list ofOrderedDictobjects are passed in (GH13304)pd.read_html()has gained support for thedecimaloption (GH12907)Serieshas gained the properties.is_monotonic,.is_monotonic_increasing,.is_monotonic_decreasing, similar toIndex(GH13336)DataFrame.to_sql()now allows a single value as the SQL type for all columns (GH11886).Series.appendnow supports theignore_indexoption (GH13677).to_stata()andStataWritercan now write variable labels to Stata dta files using a dictionary to make column names to labels (GH13535, GH13536).to_stata()andStataWriterwill automatically convertdatetime64[ns]columns to Stata format%tc, rather than raising aValueError(GH12259)read_stata()andStataReaderraise with a more explicit error message when reading Stata files with repeated value labels whenconvert_categoricals=True(GH13923)DataFrame.stylewill now render sparsified MultiIndexes (GH11655)DataFrame.stylewill now show column level names (e.g.DataFrame.columns.names) (GH13775)DataFramehas gained support to re-order the columns based on the values in a row usingdf.sort_values(by='...', axis=1)(GH10806)In [75]: df = pd.DataFrame({'A': [2, 7], 'B': [3, 5], 'C': [4, 8]}, ....: index=['row1', 'row2']) ....: In [76]: df Out[76]: A B C row1 2 3 4 row2 7 5 8 In [77]: df.sort_values(by='row2', axis=1) ����������������������������������������������������Out[77]: B A C row1 3 2 4 row2 5 7 8
Added documentation to I/O regarding the perils of reading in columns with mixed dtypes and how to handle it (GH13746)
to_html()now has aborderargument to control the value in the opening<table>tag. The default is the value of thehtml.borderoption, which defaults to 1. This also affects the notebook HTML repr, but since Jupyter’s CSS includes a border-width attribute, the visual effect is the same. (GH11563).Raise
ImportErrorin the sql functions whensqlalchemyis not installed and a connection string is used (GH11920).Compatibility with matplotlib 2.0. Older versions of pandas should also work with matplotlib 2.0 (GH13333)
Timestamp,Period,DatetimeIndex,PeriodIndexand.dtaccessor have gained a.is_leap_yearproperty to check whether the date belongs to a leap year. (GH13727)astype()will now accept a dict of column name to data types mapping as thedtypeargument. (GH12086)The
pd.read_jsonandDataFrame.to_jsonhas gained support for reading and writing json lines withlinesoption see Line delimited json (GH9180)read_excel()now supports the true_values and false_values keyword arguments (GH13347)groupby()will now accept a scalar and a single-element list for specifyinglevelon a non-MultiIndexgrouper. (GH13907)Non-convertible dates in an excel date column will be returned without conversion and the column will be
objectdtype, rather than raising an exception (GH10001).pd.Timedelta(None)is now accepted and will returnNaT, mirroringpd.Timestamp(GH13687)pd.read_stata()can now handle some format 111 files, which are produced by SAS when generating Stata dta files (GH11526)SeriesandIndexnow supportdivmodwhich will return a tuple of series or indices. This behaves like a standard binary operator with regards to broadcasting rules (GH14208).
API changes¶
Series.tolist() will now return Python types¶
Series.tolist() will now return Python types in the output, mimicking NumPy .tolist() behavior (GH10904)
In [78]: s = pd.Series([1,2,3])
Previous behavior:
In [7]: type(s.tolist()[0])
Out[7]:
<class 'numpy.int64'>
New behavior:
In [79]: type(s.tolist()[0])
Out[79]: int
Series operators for different indexes¶
Following Series operators have been changed to make all operators consistent,
including DataFrame (GH1134, GH4581, GH13538)
Seriescomparison operators now raiseValueErrorwhenindexare different.Serieslogical operators align bothindexof left and right hand side.
Warning
Until 0.18.1, comparing Series with the same length, would succeed even if
the .index are different (the result ignores .index). As of 0.19.0, this will raises ValueError to be more strict. This section also describes how to keep previous behavior or align different indexes, using the flexible comparison methods like .eq.
As a result, Series and DataFrame operators behave as below:
Arithmetic operators¶
Arithmetic operators align both index (no changes).
In [80]: s1 = pd.Series([1, 2, 3], index=list('ABC'))
In [81]: s2 = pd.Series([2, 2, 2], index=list('ABD'))
In [82]: s1 + s2
Out[82]:
A 3.0
B 4.0
C NaN
D NaN
dtype: float64
In [83]: df1 = pd.DataFrame([1, 2, 3], index=list('ABC'))
In [84]: df2 = pd.DataFrame([2, 2, 2], index=list('ABD'))
In [85]: df1 + df2
Out[85]:
0
A 3.0
B 4.0
C NaN
D NaN
Comparison operators¶
Comparison operators raise ValueError when .index are different.
Previous Behavior (Series):
Series compared values ignoring the .index as long as both had the same length:
In [1]: s1 == s2
Out[1]:
A False
B True
C False
dtype: bool
New behavior (Series):
In [2]: s1 == s2
Out[2]:
ValueError: Can only compare identically-labeled Series objects
Note
To achieve the same result as previous versions (compare values based on locations ignoring .index), compare both .values.
In [86]: s1.values == s2.values
Out[86]: array([False, True, False], dtype=bool)
If you want to compare Series aligning its .index, see flexible comparison methods section below:
In [87]: s1.eq(s2)
Out[87]:
A False
B True
C False
D False
dtype: bool
Current Behavior (DataFrame, no change):
In [3]: df1 == df2
Out[3]:
ValueError: Can only compare identically-labeled DataFrame objects
Logical operators¶
Logical operators align both .index of left and right hand side.
Previous behavior (Series), only left hand side index was kept:
In [4]: s1 = pd.Series([True, False, True], index=list('ABC'))
In [5]: s2 = pd.Series([True, True, True], index=list('ABD'))
In [6]: s1 & s2
Out[6]:
A True
B False
C False
dtype: bool
New behavior (Series):
In [88]: s1 = pd.Series([True, False, True], index=list('ABC'))
In [89]: s2 = pd.Series([True, True, True], index=list('ABD'))
In [90]: s1 & s2
Out[90]:
A True
B False
C False
D False
dtype: bool
Note
Series logical operators fill a NaN result with False.
Note
To achieve the same result as previous versions (compare values based on only left hand side index), you can use reindex_like:
In [91]: s1 & s2.reindex_like(s1)
Out[91]:
A True
B False
C False
dtype: bool
Current Behavior (DataFrame, no change):
In [92]: df1 = pd.DataFrame([True, False, True], index=list('ABC'))
In [93]: df2 = pd.DataFrame([True, True, True], index=list('ABD'))
In [94]: df1 & df2
Out[94]:
0
A True
B False
C NaN
D NaN
Flexible comparison methods¶
Series flexible comparison methods like eq, ne, le, lt, ge and gt now align both index. Use these operators if you want to compare two Series
which has the different index.
In [95]: s1 = pd.Series([1, 2, 3], index=['a', 'b', 'c'])
In [96]: s2 = pd.Series([2, 2, 2], index=['b', 'c', 'd'])
In [97]: s1.eq(s2)
Out[97]:
a False
b True
c False
d False
dtype: bool
In [98]: s1.ge(s2)
������������������������������������������������������������������Out[98]:
a False
b True
c True
d False
dtype: bool
Previously, this worked the same as comparison operators (see above).
Series type promotion on assignment¶
A Series will now correctly promote its dtype for assignment with incompat values to the current dtype (GH13234)
In [99]: s = pd.Series()
Previous behavior:
In [2]: s["a"] = pd.Timestamp("2016-01-01")
In [3]: s["b"] = 3.0
TypeError: invalid type promotion
New behavior:
In [100]: s["a"] = pd.Timestamp("2016-01-01")
In [101]: s["b"] = 3.0
In [102]: s
Out[102]:
a 2016-01-01 00:00:00
b 3
dtype: object
In [103]: s.dtype
���������������������������������������������������������������������������Out[103]: dtype('O')
.to_datetime() changes¶
Previously if .to_datetime() encountered mixed integers/floats and strings, but no datetimes with errors='coerce' it would convert all to NaT.
Previous behavior:
In [2]: pd.to_datetime([1, 'foo'], errors='coerce')
Out[2]: DatetimeIndex(['NaT', 'NaT'], dtype='datetime64[ns]', freq=None)
Current behavior:
This will now convert integers/floats with the default unit of ns.
In [104]: pd.to_datetime([1, 'foo'], errors='coerce')
Out[104]: DatetimeIndex(['1970-01-01 00:00:00.000000001', 'NaT'], dtype='datetime64[ns]', freq=None)
Bug fixes related to .to_datetime():
- Bug in
pd.to_datetime()when passing integers or floats, and nounitanderrors='coerce'(GH13180). - Bug in
pd.to_datetime()when passing invalid datatypes (e.g. bool); will now respect theerrorskeyword (GH13176) - Bug in
pd.to_datetime()which overflowed onint8, andint16dtypes (GH13451) - Bug in
pd.to_datetime()raiseAttributeErrorwithNaNand the other string is not valid whenerrors='ignore'(GH12424) - Bug in
pd.to_datetime()did not cast floats correctly whenunitwas specified, resulting in truncated datetime (GH13834)
Merging changes¶
Merging will now preserve the dtype of the join keys (GH8596)
In [105]: df1 = pd.DataFrame({'key': [1], 'v1': [10]})
In [106]: df1
Out[106]:
key v1
0 1 10
In [107]: df2 = pd.DataFrame({'key': [1, 2], 'v1': [20, 30]})
In [108]: df2
Out[108]:
key v1
0 1 20
1 2 30
Previous behavior:
In [5]: pd.merge(df1, df2, how='outer')
Out[5]:
key v1
0 1.0 10.0
1 1.0 20.0
2 2.0 30.0
In [6]: pd.merge(df1, df2, how='outer').dtypes
Out[6]:
key float64
v1 float64
dtype: object
New behavior:
We are able to preserve the join keys
In [109]: pd.merge(df1, df2, how='outer')
Out[109]:
key v1
0 1 10
1 1 20
2 2 30
In [110]: pd.merge(df1, df2, how='outer').dtypes
�������������������������������������������������������Out[110]:
key int64
v1 int64
dtype: object
Of course if you have missing values that are introduced, then the resulting dtype will be upcast, which is unchanged from previous.
In [111]: pd.merge(df1, df2, how='outer', on='key')
Out[111]:
key v1_x v1_y
0 1 10.0 20
1 2 NaN 30
In [112]: pd.merge(df1, df2, how='outer', on='key').dtypes
��������������������������������������������������������������������Out[112]:
key int64
v1_x float64
v1_y int64
dtype: object
.describe() changes¶
Percentile identifiers in the index of a .describe() output will now be rounded to the least precision that keeps them distinct (GH13104)
In [113]: s = pd.Series([0, 1, 2, 3, 4])
In [114]: df = pd.DataFrame([0, 1, 2, 3, 4])
Previous behavior:
The percentiles were rounded to at most one decimal place, which could raise ValueError for a data frame if the percentiles were duplicated.
In [3]: s.describe(percentiles=[0.0001, 0.0005, 0.001, 0.999, 0.9995, 0.9999])
Out[3]:
count 5.000000
mean 2.000000
std 1.581139
min 0.000000
0.0% 0.000400
0.1% 0.002000
0.1% 0.004000
50% 2.000000
99.9% 3.996000
100.0% 3.998000
100.0% 3.999600
max 4.000000
dtype: float64
In [4]: df.describe(percentiles=[0.0001, 0.0005, 0.001, 0.999, 0.9995, 0.9999])
Out[4]:
...
ValueError: cannot reindex from a duplicate axis
New behavior:
In [115]: s.describe(percentiles=[0.0001, 0.0005, 0.001, 0.999, 0.9995, 0.9999])
Out[115]:
count 5.000000
mean 2.000000
std 1.581139
min 0.000000
0.01% 0.000400
0.05% 0.002000
0.1% 0.004000
50% 2.000000
99.9% 3.996000
99.95% 3.998000
99.99% 3.999600
max 4.000000
dtype: float64
In [116]: df.describe(percentiles=[0.0001, 0.0005, 0.001, 0.999, 0.9995, 0.9999])
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[116]:
0
count 5.000000
mean 2.000000
std 1.581139
min 0.000000
0.01% 0.000400
0.05% 0.002000
0.1% 0.004000
50% 2.000000
99.9% 3.996000
99.95% 3.998000
99.99% 3.999600
max 4.000000
Furthermore:
- Passing duplicated
percentileswill now raise aValueError. - Bug in
.describe()on a DataFrame with a mixed-dtype column index, which would previously raise aTypeError(GH13288)
Period changes¶
PeriodIndex now has period dtype¶
PeriodIndex now has its own period dtype. The period dtype is a
pandas extension dtype like category or the timezone aware dtype (datetime64[ns, tz]) (GH13941).
As a consequence of this change, PeriodIndex no longer has an integer dtype:
Previous behavior:
In [1]: pi = pd.PeriodIndex(['2016-08-01'], freq='D')
In [2]: pi
Out[2]: PeriodIndex(['2016-08-01'], dtype='int64', freq='D')
In [3]: pd.api.types.is_integer_dtype(pi)
Out[3]: True
In [4]: pi.dtype
Out[4]: dtype('int64')
New behavior:
In [117]: pi = pd.PeriodIndex(['2016-08-01'], freq='D')
In [118]: pi
Out[118]: PeriodIndex(['2016-08-01'], dtype='period[D]', freq='D')
In [119]: pd.api.types.is_integer_dtype(pi)
�������������������������������������������������������������������Out[119]: False
In [120]: pd.api.types.is_period_dtype(pi)
�����������������������������������������������������������������������������������Out[120]: True
In [121]: pi.dtype
��������������������������������������������������������������������������������������������������Out[121]: period[D]
In [122]: type(pi.dtype)
����������������������������������������������������������������������������������������������������������������������Out[122]: pandas.core.dtypes.dtypes.PeriodDtype
Period('NaT') now returns pd.NaT¶
Previously, Period has its own Period('NaT') representation different from pd.NaT. Now Period('NaT') has been changed to return pd.NaT. (GH12759, GH13582)
Previous behavior:
In [5]: pd.Period('NaT', freq='D')
Out[5]: Period('NaT', 'D')
New behavior:
These result in pd.NaT without providing freq option.
In [123]: pd.Period('NaT')
Out[123]: NaT
In [124]: pd.Period(None)
��������������Out[124]: NaT
To be compatible with Period addition and subtraction, pd.NaT now supports addition and subtraction with int. Previously it raised ValueError.
Previous behavior:
In [5]: pd.NaT + 1
...
ValueError: Cannot add integral value to Timestamp without freq.
New behavior:
In [125]: pd.NaT + 1
Out[125]: NaT
In [126]: pd.NaT - 1
��������������Out[126]: NaT
PeriodIndex.values now returns array of Period object¶
.values is changed to return an array of Period objects, rather than an array
of integers (GH13988).
Previous behavior:
In [6]: pi = pd.PeriodIndex(['2011-01', '2011-02'], freq='M')
In [7]: pi.values
array([492, 493])
New behavior:
In [127]: pi = pd.PeriodIndex(['2011-01', '2011-02'], freq='M')
In [128]: pi.values
Out[128]: array([Period('2011-01', 'M'), Period('2011-02', 'M')], dtype=object)
Index + / - no longer used for set operations¶
Addition and subtraction of the base Index type and of DatetimeIndex
(not the numeric index types)
previously performed set operations (set union and difference). This
behavior was already deprecated since 0.15.0 (in favor using the specific
.union() and .difference() methods), and is now disabled. When
possible, + and - are now used for element-wise operations, for
example for concatenating strings or subtracting datetimes
(GH8227, GH14127).
Previous behavior:
In [1]: pd.Index(['a', 'b']) + pd.Index(['a', 'c'])
FutureWarning: using '+' to provide set union with Indexes is deprecated, use '|' or .union()
Out[1]: Index(['a', 'b', 'c'], dtype='object')
New behavior: the same operation will now perform element-wise addition:
In [129]: pd.Index(['a', 'b']) + pd.Index(['a', 'c'])
Out[129]: Index(['aa', 'bc'], dtype='object')
Note that numeric Index objects already performed element-wise operations.
For example, the behavior of adding two integer Indexes is unchanged.
The base Index is now made consistent with this behavior.
In [130]: pd.Index([1, 2, 3]) + pd.Index([2, 3, 4])
Out[130]: Int64Index([3, 5, 7], dtype='int64')
Further, because of this change, it is now possible to subtract two DatetimeIndex objects resulting in a TimedeltaIndex:
Previous behavior:
In [1]: pd.DatetimeIndex(['2016-01-01', '2016-01-02']) - pd.DatetimeIndex(['2016-01-02', '2016-01-03'])
FutureWarning: using '-' to provide set differences with datetimelike Indexes is deprecated, use .difference()
Out[1]: DatetimeIndex(['2016-01-01'], dtype='datetime64[ns]', freq=None)
New behavior:
In [131]: pd.DatetimeIndex(['2016-01-01', '2016-01-02']) - pd.DatetimeIndex(['2016-01-02', '2016-01-03'])
Out[131]: TimedeltaIndex(['-1 days', '-1 days'], dtype='timedelta64[ns]', freq=None)
Index.difference and .symmetric_difference changes¶
Index.difference and Index.symmetric_difference will now, more consistently, treat NaN values as any other values. (GH13514)
In [132]: idx1 = pd.Index([1, 2, 3, np.nan])
In [133]: idx2 = pd.Index([0, 1, np.nan])
Previous behavior:
In [3]: idx1.difference(idx2)
Out[3]: Float64Index([nan, 2.0, 3.0], dtype='float64')
In [4]: idx1.symmetric_difference(idx2)
Out[4]: Float64Index([0.0, nan, 2.0, 3.0], dtype='float64')
New behavior:
In [134]: idx1.difference(idx2)
Out[134]: Float64Index([2.0, 3.0], dtype='float64')
In [135]: idx1.symmetric_difference(idx2)
����������������������������������������������������Out[135]: Float64Index([0.0, 2.0, 3.0], dtype='float64')
Index.unique consistently returns Index¶
Index.unique() now returns unique values as an
Index of the appropriate dtype. (GH13395).
Previously, most Index classes returned np.ndarray, and DatetimeIndex,
TimedeltaIndex and PeriodIndex returned Index to keep metadata like timezone.
Previous behavior:
In [1]: pd.Index([1, 2, 3]).unique()
Out[1]: array([1, 2, 3])
In [2]: pd.DatetimeIndex(['2011-01-01', '2011-01-02', '2011-01-03'], tz='Asia/Tokyo').unique()
Out[2]:
DatetimeIndex(['2011-01-01 00:00:00+09:00', '2011-01-02 00:00:00+09:00',
'2011-01-03 00:00:00+09:00'],
dtype='datetime64[ns, Asia/Tokyo]', freq=None)
New behavior:
In [136]: pd.Index([1, 2, 3]).unique()
Out[136]: Int64Index([1, 2, 3], dtype='int64')
In [137]: pd.DatetimeIndex(['2011-01-01', '2011-01-02', '2011-01-03'], tz='Asia/Tokyo').unique()
�����������������������������������������������Out[137]:
DatetimeIndex(['2011-01-01 00:00:00+09:00', '2011-01-02 00:00:00+09:00',
'2011-01-03 00:00:00+09:00'],
dtype='datetime64[ns, Asia/Tokyo]', freq=None)
MultiIndex constructors, groupby and set_index preserve categorical dtypes¶
MultiIndex.from_arrays and MultiIndex.from_product will now preserve categorical dtype
in MultiIndex levels (GH13743, GH13854).
In [138]: cat = pd.Categorical(['a', 'b'], categories=list("bac"))
In [139]: lvl1 = ['foo', 'bar']
In [140]: midx = pd.MultiIndex.from_arrays([cat, lvl1])
In [141]: midx
Out[141]:
MultiIndex(levels=[['b', 'a', 'c'], ['bar', 'foo']],
labels=[[1, 0], [1, 0]])
Previous behavior:
In [4]: midx.levels[0]
Out[4]: Index(['b', 'a', 'c'], dtype='object')
In [5]: midx.get_level_values[0]
Out[5]: Index(['a', 'b'], dtype='object')
New behavior: the single level is now a CategoricalIndex:
In [142]: midx.levels[0]
Out[142]: CategoricalIndex(['b', 'a', 'c'], categories=['b', 'a', 'c'], ordered=False, dtype='category')
In [143]: midx.get_level_values(0)
���������������������������������������������������������������������������������������������������������Out[143]: CategoricalIndex(['a', 'b'], categories=['b', 'a', 'c'], ordered=False, dtype='category')
An analogous change has been made to MultiIndex.from_product.
As a consequence, groupby and set_index also preserve categorical dtypes in indexes
In [144]: df = pd.DataFrame({'A': [0, 1], 'B': [10, 11], 'C': cat})
In [145]: df_grouped = df.groupby(by=['A', 'C']).first()
In [146]: df_set_idx = df.set_index(['A', 'C'])
Previous behavior:
In [11]: df_grouped.index.levels[1]
Out[11]: Index(['b', 'a', 'c'], dtype='object', name='C')
In [12]: df_grouped.reset_index().dtypes
Out[12]:
A int64
C object
B float64
dtype: object
In [13]: df_set_idx.index.levels[1]
Out[13]: Index(['b', 'a', 'c'], dtype='object', name='C')
In [14]: df_set_idx.reset_index().dtypes
Out[14]:
A int64
C object
B int64
dtype: object
New behavior:
In [147]: df_grouped.index.levels[1]
Out[147]: CategoricalIndex(['b', 'a', 'c'], categories=['b', 'a', 'c'], ordered=False, name='C', dtype='category')
In [148]: df_grouped.reset_index().dtypes
�������������������������������������������������������������������������������������������������������������������Out[148]:
A int64
C category
B float64
dtype: object
In [149]: df_set_idx.index.levels[1]
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[149]: CategoricalIndex(['b', 'a', 'c'], categories=['b', 'a', 'c'], ordered=False, name='C', dtype='category')
In [150]: df_set_idx.reset_index().dtypes
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[150]:
A int64
C category
B int64
dtype: object
read_csv will progressively enumerate chunks¶
When read_csv() is called with chunksize=n and without specifying an index,
each chunk used to have an independently generated index from 0 to n-1.
They are now given instead a progressive index, starting from 0 for the first chunk,
from n for the second, and so on, so that, when concatenated, they are identical to
the result of calling read_csv() without the chunksize= argument
(GH12185).
In [151]: data = 'A,B\n0,1\n2,3\n4,5\n6,7'
Previous behavior:
In [2]: pd.concat(pd.read_csv(StringIO(data), chunksize=2))
Out[2]:
A B
0 0 1
1 2 3
0 4 5
1 6 7
New behavior:
In [152]: pd.concat(pd.read_csv(StringIO(data), chunksize=2))
Out[152]:
A B
0 0 1
1 2 3
2 4 5
3 6 7
Sparse Changes¶
These changes allow pandas to handle sparse data with more dtypes, and for work to make a smoother experience with data handling.
int64 and bool support enhancements¶
Sparse data structures now gained enhanced support of int64 and bool dtype (GH667, GH13849).
Previously, sparse data were float64 dtype by default, even if all inputs were of int or bool dtype. You had to specify dtype explicitly to create sparse data with int64 dtype. Also, fill_value had to be specified explicitly because the default was np.nan which doesn’t appear in int64 or bool data.
In [1]: pd.SparseArray([1, 2, 0, 0])
Out[1]:
[1.0, 2.0, 0.0, 0.0]
Fill: nan
IntIndex
Indices: array([0, 1, 2, 3], dtype=int32)
# specifying int64 dtype, but all values are stored in sp_values because
# fill_value default is np.nan
In [2]: pd.SparseArray([1, 2, 0, 0], dtype=np.int64)
Out[2]:
[1, 2, 0, 0]
Fill: nan
IntIndex
Indices: array([0, 1, 2, 3], dtype=int32)
In [3]: pd.SparseArray([1, 2, 0, 0], dtype=np.int64, fill_value=0)
Out[3]:
[1, 2, 0, 0]
Fill: 0
IntIndex
Indices: array([0, 1], dtype=int32)
As of v0.19.0, sparse data keeps the input dtype, and uses more appropriate fill_value defaults (0 for int64 dtype, False for bool dtype).
In [153]: pd.SparseArray([1, 2, 0, 0], dtype=np.int64)
Out[153]:
[1, 2, 0, 0]
Fill: 0
IntIndex
Indices: array([0, 1], dtype=int32)
In [154]: pd.SparseArray([True, False, False, False])
�����������������������������������������������������������������������������Out[154]:
[True, False, False, False]
Fill: False
IntIndex
Indices: array([0], dtype=int32)
See the docs for more details.
Operators now preserve dtypes¶
Sparse data structure now can preserve
dtypeafter arithmetic ops (GH13848)In [155]: s = pd.SparseSeries([0, 2, 0, 1], fill_value=0, dtype=np.int64) In [156]: s.dtype Out[156]: dtype('int64') In [157]: s + 1 �������������������������Out[157]: 0 1 1 3 2 1 3 2 dtype: int64 BlockIndex Block locations: array([1, 3], dtype=int32) Block lengths: array([1, 1], dtype=int32)
Sparse data structure now support
astypeto convert internaldtype(GH13900)In [158]: s = pd.SparseSeries([1., 0., 2., 0.], fill_value=0) In [159]: s Out[159]: 0 1.0 1 0.0 2 2.0 3 0.0 dtype: float64 BlockIndex Block locations: array([0, 2], dtype=int32) Block lengths: array([1, 1], dtype=int32) In [160]: s.astype(np.int64) ���������������������������������������������������������������������������������������������������������������������������������������������������������������Out[160]: 0 1 1 0 2 2 3 0 dtype: int64 BlockIndex Block locations: array([0, 2], dtype=int32) Block lengths: array([1, 1], dtype=int32)
astypefails if data contains values which cannot be converted to specifieddtype. Note that the limitation is applied tofill_valuewhich default isnp.nan.In [7]: pd.SparseSeries([1., np.nan, 2., np.nan], fill_value=np.nan).astype(np.int64) Out[7]: ValueError: unable to coerce current fill_value nan to int64 dtype
Other sparse fixes¶
- Subclassed
SparseDataFrameandSparseSeriesnow preserve class types when slicing or transposing. (GH13787) SparseArraywithbooldtype now supports logical (bool) operators (GH14000)- Bug in
SparseSerieswithMultiIndex[]indexing may raiseIndexError(GH13144) - Bug in
SparseSerieswithMultiIndex[]indexing result may have normalIndex(GH13144) - Bug in
SparseDataFramein whichaxis=Nonedid not default toaxis=0(GH13048) - Bug in
SparseSeriesandSparseDataFramecreation withobjectdtype may raiseTypeError(GH11633) - Bug in
SparseDataFramedoesn’t respect passedSparseArrayorSparseSeries‘s dtype andfill_value(GH13866) - Bug in
SparseArrayandSparseSeriesdon’t apply ufunc tofill_value(GH13853) - Bug in
SparseSeries.absincorrectly keeps negativefill_value(GH13853) - Bug in single row slicing on multi-type
SparseDataFrames, types were previously forced to float (GH13917) - Bug in
SparseSeriesslicing changes integer dtype to float (GH8292) - Bug in
SparseDataFarmecomparison ops may raiseTypeError(GH13001) - Bug in
SparseDataFarme.isnullraisesValueError(GH8276) - Bug in
SparseSeriesrepresentation withbooldtype may raiseIndexError(GH13110) - Bug in
SparseSeriesandSparseDataFrameofboolorint64dtype may display its values likefloat64dtype (GH13110) - Bug in sparse indexing using
SparseArraywithbooldtype may return incorrect result (GH13985) - Bug in
SparseArraycreated fromSparseSeriesmay losedtype(GH13999) - Bug in
SparseSeriescomparison with dense returns normalSeriesrather thanSparseSeries(GH13999)
Indexer dtype changes¶
Note
This change only affects 64 bit python running on Windows, and only affects relatively advanced indexing operations
Methods such as Index.get_indexer that return an indexer array, coerce that array to a “platform int”, so that it can be
directly used in 3rd party library operations like numpy.take. Previously, a platform int was defined as np.int_
which corresponds to a C integer, but the correct type, and what is being used now, is np.intp, which corresponds
to the C integer size that can hold a pointer (GH3033, GH13972).
These types are the same on many platform, but for 64 bit python on Windows,
np.int_ is 32 bits, and np.intp is 64 bits. Changing this behavior improves performance for many
operations on that platform.
Previous behavior:
In [1]: i = pd.Index(['a', 'b', 'c'])
In [2]: i.get_indexer(['b', 'b', 'c']).dtype
Out[2]: dtype('int32')
New behavior:
In [1]: i = pd.Index(['a', 'b', 'c'])
In [2]: i.get_indexer(['b', 'b', 'c']).dtype
Out[2]: dtype('int64')
Other API Changes¶
Timestamp.to_pydatetimewill issue aUserWarningwhenwarn=True, and the instance has a non-zero number of nanoseconds, previously this would print a message to stdout (GH14101).Series.unique()with datetime and timezone now returns return array ofTimestampwith timezone (GH13565).Panel.to_sparse()will raise aNotImplementedErrorexception when called (GH13778).Index.reshape()will raise aNotImplementedErrorexception when called (GH12882)..filter()enforces mutual exclusion of the keyword arguments (GH12399).eval‘s upcasting rules forfloat32types have been updated to be more consistent with NumPy’s rules. New behavior will not upcast tofloat64if you multiply a pandasfloat32object by a scalar float64 (GH12388).- An
UnsupportedFunctionCallerror is now raised if NumPy ufuncs likenp.meanare called on groupby or resample objects (GH12811). __setitem__will no longer apply a callable rhs as a function instead of storing it. Callwheredirectly to get the previous behavior (GH13299).- Calls to
.sample()will respect the random seed set vianumpy.random.seed(n)(GH13161) Styler.applyis now more strict about the outputs your function must return. Foraxis=0oraxis=1, the output shape must be identical. Foraxis=None, the output must be a DataFrame with identical columns and index labels (GH13222).Float64Index.astype(int)will now raiseValueErrorifFloat64IndexcontainsNaNvalues (GH13149)TimedeltaIndex.astype(int)andDatetimeIndex.astype(int)will now returnInt64Indexinstead ofnp.array(GH13209)- Passing
Periodwith multiple frequencies to normalIndexnow returnsIndexwithobjectdtype (GH13664) PeriodIndex.fillnawithPeriodhas different freq now coerces toobjectdtype (GH13664)- Faceted boxplots from
DataFrame.boxplot(by=col)now return aSerieswhenreturn_typeis not None. Previously these returned anOrderedDict. Note that whenreturn_type=None, the default, these still return a 2-D NumPy array (GH12216, GH7096). pd.read_hdfwill now raise aValueErrorinstead ofKeyError, if a mode other thanr,r+andais supplied. (GH13623)pd.read_csv(),pd.read_table(), andpd.read_hdf()raise the builtinFileNotFoundErrorexception for Python 3.x when called on a nonexistent file; this is back-ported asIOErrorin Python 2.x (GH14086)- More informative exceptions are passed through the csv parser. The exception type would now be the original exception type instead of
CParserError(GH13652). pd.read_csv()in the C engine will now issue aParserWarningor raise aValueErrorwhensepencoded is more than one character long (GH14065)DataFrame.valueswill now returnfloat64with aDataFrameof mixedint64anduint64dtypes, conforming tonp.find_common_type(GH10364, GH13917).groupby.groupswill now return a dictionary ofIndexobjects, rather than a dictionary ofnp.ndarrayorlists(GH14293)
Deprecations¶
Series.reshapeandCategorical.reshapehave been deprecated and will be removed in a subsequent release (GH12882, GH12882)PeriodIndex.to_datetimehas been deprecated in favor ofPeriodIndex.to_timestamp(GH8254)Timestamp.to_datetimehas been deprecated in favor ofTimestamp.to_pydatetime(GH8254)Index.to_datetimeandDatetimeIndex.to_datetimehave been deprecated in favor ofpd.to_datetime(GH8254)pandas.core.datetoolsmodule has been deprecated and will be removed in a subsequent release (GH14094)SparseListhas been deprecated and will be removed in a future version (GH13784)DataFrame.to_html()andDataFrame.to_latex()have dropped thecolSpaceparameter in favor ofcol_space(GH13857)DataFrame.to_sql()has deprecated theflavorparameter, as it is superfluous when SQLAlchemy is not installed (GH13611)- Deprecated
read_csvkeywords:compact_intsanduse_unsignedhave been deprecated and will be removed in a future version (GH13320)buffer_lineshas been deprecated and will be removed in a future version (GH13360)as_recarrayhas been deprecated and will be removed in a future version (GH13373)skip_footerhas been deprecated in favor ofskipfooterand will be removed in a future version (GH13349)
- top-level
pd.ordered_merge()has been renamed topd.merge_ordered()and the original name will be removed in a future version (GH13358) Timestamp.offsetproperty (and named arg in the constructor), has been deprecated in favor offreq(GH12160)pd.tseries.util.pivot_annualis deprecated. Usepivot_tableas alternative, an example is here (GH736)pd.tseries.util.isleapyearhas been deprecated and will be removed in a subsequent release. Datetime-likes now have a.is_leap_yearproperty (GH13727)Panel4DandPanelNDconstructors are deprecated and will be removed in a future version. The recommended way to represent these types of n-dimensional data are with the xarray package. Pandas provides ato_xarray()method to automate this conversion (GH13564).pandas.tseries.frequencies.get_standard_freqis deprecated. Usepandas.tseries.frequencies.to_offset(freq).rule_codeinstead (GH13874)pandas.tseries.frequencies.to_offset‘sfreqstrkeyword is deprecated in favor offreq(GH13874)Categorical.from_arrayhas been deprecated and will be removed in a future version (GH13854)
Removal of prior version deprecations/changes¶
- The
SparsePanelclass has been removed (GH13778) - The
pd.sandboxmodule has been removed in favor of the external librarypandas-qt(GH13670) - The
pandas.io.dataandpandas.io.wbmodules are removed in favor of the pandas-datareader package (GH13724). - The
pandas.tools.rplotmodule has been removed in favor of the seaborn package (GH13855) DataFrame.to_csv()has dropped theengineparameter, as was deprecated in 0.17.1 (GH11274, GH13419)DataFrame.to_dict()has dropped theouttypeparameter in favor oforient(GH13627, GH8486)pd.Categoricalhas dropped setting of theorderedattribute directly in favor of theset_orderedmethod (GH13671)pd.Categoricalhas dropped thelevelsattribute in favor ofcategories(GH8376)DataFrame.to_sql()has dropped themysqloption for theflavorparameter (GH13611)Panel.shift()has dropped thelagsparameter in favor ofperiods(GH14041)pd.Indexhas dropped thediffmethod in favor ofdifference(GH13669)pd.DataFramehas dropped theto_widemethod in favor ofto_panel(GH14039)Series.to_csvhas dropped thenanRepparameter in favor ofna_rep(GH13804)Series.xs,DataFrame.xs,Panel.xs,Panel.major_xs, andPanel.minor_xshave dropped thecopyparameter (GH13781)str.splithas dropped thereturn_typeparameter in favor ofexpand(GH13701)- Removal of the legacy time rules (offset aliases), deprecated since 0.17.0 (this has been alias since 0.8.0) (GH13590, GH13868). Now legacy time rules raises
ValueError. For the list of currently supported offsets, see here. - The default value for the
return_typeparameter forDataFrame.plot.boxandDataFrame.boxplotchanged fromNoneto"axes". These methods will now return a matplotlib axes by default instead of a dictionary of artists. See here (GH6581). - The
tqueryanduqueryfunctions in thepandas.io.sqlmodule are removed (GH5950).
Performance Improvements¶
- Improved performance of sparse
IntIndex.intersect(GH13082) - Improved performance of sparse arithmetic with
BlockIndexwhen the number of blocks are large, though recommended to useIntIndexin such cases (GH13082) - Improved performance of
DataFrame.quantile()as it now operates per-block (GH11623) - Improved performance of float64 hash table operations, fixing some very slow indexing and groupby operations in python 3 (GH13166, GH13334)
- Improved performance of
DataFrameGroupBy.transform(GH12737) - Improved performance of
IndexandSeries.duplicated(GH10235) - Improved performance of
Index.difference(GH12044) - Improved performance of
RangeIndex.is_monotonic_increasingandis_monotonic_decreasing(GH13749) - Improved performance of datetime string parsing in
DatetimeIndex(GH13692) - Improved performance of hashing
Period(GH12817) - Improved performance of
factorizeof datetime with timezone (GH13750) - Improved performance of by lazily creating indexing hashtables on larger Indexes (GH14266)
- Improved performance of
groupby.groups(GH14293) - Unecessary materializing of a MultiIndex when introspecting for memory usage (GH14308)
Bug Fixes¶
- Bug in
groupby().shift(), which could cause a segfault or corruption in rare circumstances when grouping by columns with missing values (GH13813) - Bug in
groupby().cumsum()calculatingcumprodwhenaxis=1. (GH13994) - Bug in
pd.to_timedelta()in which theerrorsparameter was not being respected (GH13613) - Bug in
io.json.json_normalize(), where non-ascii keys raised an exception (GH13213) - Bug when passing a not-default-indexed
Seriesasxerroryerrin.plot()(GH11858) - Bug in area plot draws legend incorrectly if subplot is enabled or legend is moved after plot (matplotlib 1.5.0 is required to draw area plot legend properly) (GH9161, GH13544)
- Bug in
DataFrameassignment with an object-dtypedIndexwhere the resultant column is mutable to the original object. (GH13522) - Bug in matplotlib
AutoDataFormatter; this restores the second scaled formatting and re-adds micro-second scaled formatting (GH13131) - Bug in selection from a
HDFStorewith a fixed format andstartand/orstopspecified will now return the selected range (GH8287) - Bug in
Categorical.from_codes()where an unhelpful error was raised when an invalidorderedparameter was passed in (GH14058) - Bug in
Seriesconstruction from a tuple of integers on windows not returning default dtype (int64) (GH13646) - Bug in
TimedeltaIndexaddition with a Datetime-like object where addition overflow was not being caught (GH14068) - Bug in
.groupby(..).resample(..)when the same object is called multiple times (GH13174) - Bug in
.to_records()when index name is a unicode string (GH13172) - Bug in calling
.memory_usage()on object which doesn’t implement (GH12924) - Regression in
Series.quantilewith nans (also shows up in.median()and.describe()); furthermore now names theSerieswith the quantile (GH13098, GH13146) - Bug in
SeriesGroupBy.transformwith datetime values and missing groups (GH13191) - Bug where empty
Serieswere incorrectly coerced in datetime-like numeric operations (GH13844) - Bug in
Categoricalconstructor when passed aCategoricalcontaining datetimes with timezones (GH14190) - Bug in
Series.str.extractall()withstrindex raisesValueError(GH13156) - Bug in
Series.str.extractall()with single group and quantifier (GH13382) - Bug in
DatetimeIndexandPeriodsubtraction raisesValueErrororAttributeErrorrather thanTypeError(GH13078) - Bug in
IndexandSeriescreated withNaNandNaTmixed data may not havedatetime64dtype (GH13324) - Bug in
IndexandSeriesmay ignorenp.datetime64('nat')andnp.timdelta64('nat')to infer dtype (GH13324) - Bug in
PeriodIndexandPeriodsubtraction raisesAttributeError(GH13071) - Bug in
PeriodIndexconstruction returning afloat64index in some circumstances (GH13067) - Bug in
.resample(..)with aPeriodIndexnot changing itsfreqappropriately when empty (GH13067) - Bug in
.resample(..)with aPeriodIndexnot retaining its type or name with an emptyDataFrameappropriately when empty (GH13212) - Bug in
groupby(..).apply(..)when the passed function returns scalar values per group (GH13468). - Bug in
groupby(..).resample(..)where passing some keywords would raise an exception (GH13235) - Bug in
.tz_converton a tz-awareDateTimeIndexthat relied on index being sorted for correct results (GH13306) - Bug in
.tz_localizewithdateutil.tz.tzlocalmay return incorrect result (GH13583) - Bug in
DatetimeTZDtypedtype withdateutil.tz.tzlocalcannot be regarded as valid dtype (GH13583) - Bug in
pd.read_hdf()where attempting to load an HDF file with a single dataset, that had one or more categorical columns, failed unless the key argument was set to the name of the dataset. (GH13231) - Bug in
.rolling()that allowed a negative integer window in contruction of theRolling()object, but would later fail on aggregation (GH13383) - Bug in
Seriesindexing with tuple-valued data and a numeric index (GH13509) - Bug in printing
pd.DataFramewhere unusual elements with theobjectdtype were causing segfaults (GH13717) - Bug in ranking
Serieswhich could result in segfaults (GH13445) - Bug in various index types, which did not propagate the name of passed index (GH12309)
- Bug in
DatetimeIndex, which did not honour thecopy=True(GH13205) - Bug in
DatetimeIndex.is_normalizedreturns incorrectly for normalized date_range in case of local timezones (GH13459) - Bug in
pd.concatand.appendmay coercesdatetime64andtimedeltatoobjectdtype containing python built-indatetimeortimedeltarather thanTimestamporTimedelta(GH13626) - Bug in
PeriodIndex.appendmay raisesAttributeErrorwhen the result isobjectdtype (GH13221) - Bug in
CategoricalIndex.appendmay accept normallist(GH13626) - Bug in
pd.concatand.appendwith the same timezone get reset to UTC (GH7795) - Bug in
SeriesandDataFrame.appendraisesAmbiguousTimeErrorif data contains datetime near DST boundary (GH13626) - Bug in
DataFrame.to_csv()in which float values were being quoted even though quotations were specified for non-numeric values only (GH12922, GH13259) - Bug in
DataFrame.describe()raisingValueErrorwith only boolean columns (GH13898) - Bug in
MultiIndexslicing where extra elements were returned when level is non-unique (GH12896) - Bug in
.str.replacedoes not raiseTypeErrorfor invalid replacement (GH13438) - Bug in
MultiIndex.from_arrayswhich didn’t check for input array lengths matching (GH13599) - Bug in
cartesian_productandMultiIndex.from_productwhich may raise with empty input arrays (GH12258) - Bug in
pd.read_csv()which may cause a segfault or corruption when iterating in large chunks over a stream/file under rare circumstances (GH13703) - Bug in
pd.read_csv()which caused errors to be raised when a dictionary containing scalars is passed in forna_values(GH12224) - Bug in
pd.read_csv()which caused BOM files to be incorrectly parsed by not ignoring the BOM (GH4793) - Bug in
pd.read_csv()withengine='python'which raised errors when a numpy array was passed in forusecols(GH12546) - Bug in
pd.read_csv()where the index columns were being incorrectly parsed when parsed as dates with athousandsparameter (GH14066) - Bug in
pd.read_csv()withengine='python'in whichNaNvalues weren’t being detected after data was converted to numeric values (GH13314) - Bug in
pd.read_csv()in which thenrowsargument was not properly validated for both engines (GH10476) - Bug in
pd.read_csv()withengine='python'in which infinities of mixed-case forms were not being interpreted properly (GH13274) - Bug in
pd.read_csv()withengine='python'in which trailingNaNvalues were not being parsed (GH13320) - Bug in
pd.read_csv()withengine='python'when reading from atempfile.TemporaryFileon Windows with Python 3 (GH13398) - Bug in
pd.read_csv()that preventsusecolskwarg from accepting single-byte unicode strings (GH13219) - Bug in
pd.read_csv()that preventsusecolsfrom being an empty set (GH13402) - Bug in
pd.read_csv()in the C engine where the NULL character was not being parsed as NULL (GH14012) - Bug in
pd.read_csv()withengine='c'in which NULLquotecharwas not accepted even thoughquotingwas specified asNone(GH13411) - Bug in
pd.read_csv()withengine='c'in which fields were not properly cast to float when quoting was specified as non-numeric (GH13411) - Bug in
pd.read_csv()in Python 2.x with non-UTF8 encoded, multi-character separated data (GH3404) - Bug in
pd.read_csv(), where aliases for utf-xx (e.g. UTF-xx, UTF_xx, utf_xx) raised UnicodeDecodeError (GH13549) - Bug in
pd.read_csv,pd.read_table,pd.read_fwf,pd.read_stataandpd.read_saswhere files were opened by parsers but not closed if bothchunksizeanditeratorwereNone. (GH13940) - Bug in
StataReader,StataWriter,XportReaderandSAS7BDATReaderwhere a file was not properly closed when an error was raised. (GH13940) - Bug in
pd.pivot_table()wheremargins_nameis ignored whenaggfuncis a list (GH13354) - Bug in
pd.Series.str.zfill,center,ljust,rjust, andpadwhen passing non-integers, did not raiseTypeError(GH13598) - Bug in checking for any null objects in a
TimedeltaIndex, which always returnedTrue(GH13603) - Bug in
Seriesarithmetic raisesTypeErrorif it contains datetime-like asobjectdtype (GH13043) - Bug
Series.isnull()andSeries.notnull()ignorePeriod('NaT')(GH13737) - Bug
Series.fillna()andSeries.dropna()don’t affect toPeriod('NaT')(GH13737 - Bug in
.fillna(value=np.nan)incorrectly raisesKeyErroron acategorydtypedSeries(GH14021) - Bug in extension dtype creation where the created types were not is/identical (GH13285)
- Bug in
.resample(..)where incorrect warnings were triggered by IPython introspection (GH13618) - Bug in
NaT-PeriodraisesAttributeError(GH13071) - Bug in
Seriescomparison may output incorrect result if rhs containsNaT(GH9005) - Bug in
SeriesandIndexcomparison may output incorrect result if it containsNaTwithobjectdtype (GH13592) - Bug in
Periodaddition raisesTypeErrorifPeriodis on right hand side (GH13069) - Bug in
PeirodandSeriesorIndexcomparison raisesTypeError(GH13200) - Bug in
pd.set_eng_float_format()that would prevent NaN and Inf from formatting (GH11981) - Bug in
.unstackwithCategoricaldtype resets.orderedtoTrue(GH13249) - Clean some compile time warnings in datetime parsing (GH13607)
- Bug in
factorizeraisesAmbiguousTimeErrorif data contains datetime near DST boundary (GH13750) - Bug in
.set_indexraisesAmbiguousTimeErrorif new index contains DST boundary and multi levels (GH12920) - Bug in
.shiftraisesAmbiguousTimeErrorif data contains datetime near DST boundary (GH13926) - Bug in
pd.read_hdf()returns incorrect result when aDataFramewith acategoricalcolumn and a query which doesn’t match any values (GH13792) - Bug in
.ilocwhen indexing with a non lex-sorted MultiIndex (GH13797) - Bug in
.locwhen indexing with date strings in a reverse sortedDatetimeIndex(GH14316) - Bug in
Seriescomparison operators when dealing with zero dim NumPy arrays (GH13006) - Bug in
.combine_firstmay return incorrectdtype(GH7630, GH10567) - Bug in
groupbywhereapplyreturns different result depending on whether first result isNoneor not (GH12824) - Bug in
groupby(..).nth()where the group key is included inconsistently if called after.head()/.tail()(GH12839) - Bug in
.to_html,.to_latexand.to_stringsilently ignore custom datetime formatter passed through theformatterskey word (GH10690) - Bug in
DataFrame.iterrows(), not yielding aSeriessubclasse if defined (GH13977) - Bug in
pd.to_numericwhenerrors='coerce'and input contains non-hashable objects (GH13324) - Bug in invalid
Timedeltaarithmetic and comparison may raiseValueErrorrather thanTypeError(GH13624) - Bug in invalid datetime parsing in
to_datetimeandDatetimeIndexmay raiseTypeErrorrather thanValueError(GH11169, GH11287) - Bug in
Indexcreated with tz-awareTimestampand mismatchedtzoption incorrectly coerces timezone (GH13692) - Bug in
DatetimeIndexwith nanosecond frequency does not include timestamp specified withend(GH13672) - Bug in
`Series`when setting a slice with a`np.timedelta64`(GH14155) - Bug in
IndexraisesOutOfBoundsDatetimeifdatetimeexceedsdatetime64[ns]bounds, rather than coercing toobjectdtype (GH13663) - Bug in
Indexmay ignore specifieddatetime64ortimedelta64passed asdtype(GH13981) - Bug in
RangeIndexcan be created without no arguments rather than raisesTypeError(GH13793) - Bug in
.value_counts()raisesOutOfBoundsDatetimeif data exceedsdatetime64[ns]bounds (GH13663) - Bug in
DatetimeIndexmay raiseOutOfBoundsDatetimeif inputnp.datetime64has other unit thanns(GH9114) - Bug in
Seriescreation withnp.datetime64which has other unit thannsasobjectdtype results in incorrect values (GH13876) - Bug in
resamplewith timedelta data where data was casted to float (GH13119). - Bug in
pd.isnull()pd.notnull()raiseTypeErrorif input datetime-like has other unit thanns(GH13389) - Bug in
pd.merge()may raiseTypeErrorif input datetime-like has other unit thanns(GH13389) - Bug in
HDFStore/read_hdf()discardedDatetimeIndex.nameiftzwas set (GH13884) - Bug in
Categorical.remove_unused_categories()changes.codesdtype to platform int (GH13261) - Bug in
groupbywithas_index=Falsereturns all NaN’s when grouping on multiple columns including a categorical one (GH13204) - Bug in
df.groupby(...)[...]where getitem withInt64Indexraised an error (GH13731) - Bug in the CSS classes assigned to
DataFrame.stylefor index names. Previously they were assigned"col_heading level<n> col<c>"wherenwas the number of levels + 1. Now they are assigned"index_name level<n>", wherenis the correct level for that MultiIndex. - Bug where
pd.read_gbq()could throwImportError: No module named discoveryas a result of a naming conflict with another python package called apiclient (GH13454) - Bug in
Index.unionreturns an incorrect result with a named empty index (GH13432) - Bugs in
Index.differenceandDataFrame.joinraise in Python3 when using mixed-integer indexes (GH13432, GH12814) - Bug in subtract tz-aware
datetime.datetimefrom tz-awaredatetime64series (GH14088) - Bug in
.to_excel()when DataFrame contains a MultiIndex which contains a label with a NaN value (GH13511) - Bug in invalid frequency offset string like “D1”, “-2-3H” may not raise
ValueError(GH13930) - Bug in
concatandgroupbyfor hierarchical frames withRangeIndexlevels (GH13542). - Bug in
Series.str.contains()for Series containing onlyNaNvalues ofobjectdtype (GH14171) - Bug in
agg()function on groupby dataframe changes dtype ofdatetime64[ns]column tofloat64(GH12821) - Bug in using NumPy ufunc with
PeriodIndexto add or subtract integer raiseIncompatibleFrequency. Note that using standard operator like+or-is recommended, because standard operators use more efficient path (GH13980) - Bug in operations on
NaTreturningfloatinstead ofdatetime64[ns](GH12941) - Bug in
Seriesflexible arithmetic methods (like.add()) raisesValueErrorwhenaxis=None(GH13894) - Bug in
DataFrame.to_csv()withMultiIndexcolumns in which a stray empty line was added (GH6618) - Bug in
DatetimeIndex,TimedeltaIndexandPeriodIndex.equals()may returnTruewhen input isn’tIndexbut contains the same values (GH13107) - Bug in assignment against datetime with timezone may not work if it contains datetime near DST boundary (GH14146)
- Bug in
pd.eval()andHDFStorequery truncating long float literals with python 2 (GH14241) - Bug in
IndexraisesKeyErrordisplaying incorrect column when column is not in the df and columns contains duplicate values (GH13822) - Bug in
PeriodandPeriodIndexcreating wrong dates when frequency has combined offset aliases (GH13874) - Bug in
.to_string()when called with an integerline_widthandindex=Falseraises an UnboundLocalError exception becauseidxreferenced before assignment. - Bug in
eval()where theresolversargument would not accept a list (GH14095) - Bugs in
stack,get_dummies,make_axis_dummieswhich don’t preserve categorical dtypes in (multi)indexes (GH13854) PeriodIndexcan now acceptlistandarraywhich containspd.NaT(GH13430)- Bug in
df.groupbywhere.median()returns arbitrary values if grouped dataframe contains empty bins (GH13629) - Bug in
Index.copy()wherenameparameter was ignored (GH14302)
v0.18.1 (May 3, 2016)¶
This is a minor bug-fix release from 0.18.0 and includes a large number of bug fixes along with several new features, enhancements, and performance improvements. We recommend that all users upgrade to this version.
Highlights include:
.groupby(...)has been enhanced to provide convenient syntax when working with.rolling(..),.expanding(..)and.resample(..)per group, see herepd.to_datetime()has gained the ability to assemble dates from aDataFrame, see here- Method chaining improvements, see here.
- Custom business hour offset, see here.
- Many bug fixes in the handling of
sparse, see here - Expanded the Tutorials section with a feature on modern pandas, courtesy of @TomAugsburger. (GH13045).
What’s new in v0.18.1
New features¶
Custom Business Hour¶
The CustomBusinessHour is a mixture of BusinessHour and CustomBusinessDay which
allows you to specify arbitrary holidays. For details,
see Custom Business Hour (GH11514)
In [1]: from pandas.tseries.offsets import CustomBusinessHour
In [2]: from pandas.tseries.holiday import USFederalHolidayCalendar
In [3]: bhour_us = CustomBusinessHour(calendar=USFederalHolidayCalendar())
Friday before MLK Day
In [4]: dt = datetime(2014, 1, 17, 15)
In [5]: dt + bhour_us
Out[5]: Timestamp('2014-01-17 16:00:00')
Tuesday after MLK Day (Monday is skipped because it’s a holiday)
In [6]: dt + bhour_us * 2
Out[6]: Timestamp('2014-01-20 09:00:00')
.groupby(..) syntax with window and resample operations¶
.groupby(...) has been enhanced to provide convenient syntax when working with .rolling(..), .expanding(..) and .resample(..) per group, see (GH12486, GH12738).
You can now use .rolling(..) and .expanding(..) as methods on groupbys. These return another deferred object (similar to what .rolling() and .expanding() do on ungrouped pandas objects). You can then operate on these RollingGroupby objects in a similar manner.
Previously you would have to do this to get a rolling window mean per-group:
In [7]: df = pd.DataFrame({'A': [1] * 20 + [2] * 12 + [3] * 8,
...: 'B': np.arange(40)})
...:
In [8]: df
Out[8]:
A B
0 1 0
1 1 1
2 1 2
3 1 3
4 1 4
5 1 5
6 1 6
.. .. ..
33 3 33
34 3 34
35 3 35
36 3 36
37 3 37
38 3 38
39 3 39
[40 rows x 2 columns]
In [9]: df.groupby('A').apply(lambda x: x.rolling(4).B.mean())
Out[9]:
A
1 0 NaN
1 NaN
2 NaN
3 1.5
4 2.5
5 3.5
6 4.5
...
3 33 NaN
34 NaN
35 33.5
36 34.5
37 35.5
38 36.5
39 37.5
Name: B, Length: 40, dtype: float64
Now you can do:
In [10]: df.groupby('A').rolling(4).B.mean()
Out[10]:
A
1 0 NaN
1 NaN
2 NaN
3 1.5
4 2.5
5 3.5
6 4.5
...
3 33 NaN
34 NaN
35 33.5
36 34.5
37 35.5
38 36.5
39 37.5
Name: B, Length: 40, dtype: float64
For .resample(..) type of operations, previously you would have to:
In [11]: df = pd.DataFrame({'date': pd.date_range(start='2016-01-01',
....: periods=4,
....: freq='W'),
....: 'group': [1, 1, 2, 2],
....: 'val': [5, 6, 7, 8]}).set_index('date')
....:
In [12]: df
Out[12]:
group val
date
2016-01-03 1 5
2016-01-10 1 6
2016-01-17 2 7
2016-01-24 2 8
In [13]: df.groupby('group').apply(lambda x: x.resample('1D').ffill())
Out[13]:
group val
group date
1 2016-01-03 1 5
2016-01-04 1 5
2016-01-05 1 5
2016-01-06 1 5
2016-01-07 1 5
2016-01-08 1 5
2016-01-09 1 5
... ... ...
2 2016-01-18 2 7
2016-01-19 2 7
2016-01-20 2 7
2016-01-21 2 7
2016-01-22 2 7
2016-01-23 2 7
2016-01-24 2 8
[16 rows x 2 columns]
Now you can do:
In [14]: df.groupby('group').resample('1D').ffill()
Out[14]:
group val
group date
1 2016-01-03 1 5
2016-01-04 1 5
2016-01-05 1 5
2016-01-06 1 5
2016-01-07 1 5
2016-01-08 1 5
2016-01-09 1 5
... ... ...
2 2016-01-18 2 7
2016-01-19 2 7
2016-01-20 2 7
2016-01-21 2 7
2016-01-22 2 7
2016-01-23 2 7
2016-01-24 2 8
[16 rows x 2 columns]
Method chaininng improvements¶
The following methods / indexers now accept a callable. It is intended to make
these more useful in method chains, see the documentation.
(GH11485, GH12533)
.where()and.mask().loc[],iloc[]and.ix[][]indexing
.where() and .mask()¶
These can accept a callable for the condition and other
arguments.
In [15]: df = pd.DataFrame({'A': [1, 2, 3],
....: 'B': [4, 5, 6],
....: 'C': [7, 8, 9]})
....:
In [16]: df.where(lambda x: x > 4, lambda x: x + 10)
Out[16]:
A B C
0 11 14 7
1 12 5 8
2 13 6 9
.loc[], .iloc[], .ix[]¶
These can accept a callable, and a tuple of callable as a slicer. The callable can return a valid boolean indexer or anything which is valid for these indexer’s input.
# callable returns bool indexer
In [17]: df.loc[lambda x: x.A >= 2, lambda x: x.sum() > 10]
Out[17]:
B C
1 5 8
2 6 9
# callable returns list of labels
In [18]: df.loc[lambda x: [1, 2], lambda x: ['A', 'B']]
����������������������������������Out[18]:
A B
1 2 5
2 3 6
[] indexing¶
Finally, you can use a callable in [] indexing of Series, DataFrame and Panel.
The callable must return a valid input for [] indexing depending on its
class and index type.
In [19]: df[lambda x: 'A']
Out[19]:
0 1
1 2
2 3
Name: A, dtype: int64
Using these methods / indexers, you can chain data selection operations without using temporary variable.
In [20]: bb = pd.read_csv('data/baseball.csv', index_col='id')
In [21]: (bb.groupby(['year', 'team'])
....: .sum()
....: .loc[lambda df: df.r > 100]
....: )
....:
Out[21]:
stint g ab r h X2b X3b hr rbi sb cs bb \
year team
2007 CIN 6 379 745 101 203 35 2 36 125.0 10.0 1.0 105
DET 5 301 1062 162 283 54 4 37 144.0 24.0 7.0 97
HOU 4 311 926 109 218 47 6 14 77.0 10.0 4.0 60
LAN 11 413 1021 153 293 61 3 36 154.0 7.0 5.0 114
NYN 13 622 1854 240 509 101 3 61 243.0 22.0 4.0 174
SFN 5 482 1305 198 337 67 6 40 171.0 26.0 7.0 235
TEX 2 198 729 115 200 40 4 28 115.0 21.0 4.0 73
TOR 4 459 1408 187 378 96 2 58 223.0 4.0 2.0 190
so ibb hbp sh sf gidp
year team
2007 CIN 127.0 14.0 1.0 1.0 15.0 18.0
DET 176.0 3.0 10.0 4.0 8.0 28.0
HOU 212.0 3.0 9.0 16.0 6.0 17.0
LAN 141.0 8.0 9.0 3.0 8.0 29.0
NYN 310.0 24.0 23.0 18.0 15.0 48.0
SFN 188.0 51.0 8.0 16.0 6.0 41.0
TEX 140.0 4.0 5.0 2.0 8.0 16.0
TOR 265.0 16.0 12.0 4.0 16.0 38.0
Partial string indexing on DateTimeIndex when part of a MultiIndex¶
Partial string indexing now matches on DateTimeIndex when part of a MultiIndex (GH10331)
In [22]: dft2 = pd.DataFrame(np.random.randn(20, 1),
....: columns=['A'],
....: index=pd.MultiIndex.from_product([pd.date_range('20130101',
....: periods=10,
....: freq='12H'),
....: ['a', 'b']]))
....:
In [23]: dft2
Out[23]:
A
2013-01-01 00:00:00 a 0.156998
b -0.571455
2013-01-01 12:00:00 a 1.057633
b -0.791489
2013-01-02 00:00:00 a -0.524627
b 0.071878
2013-01-02 12:00:00 a 1.910759
... ...
2013-01-04 00:00:00 b 1.015405
2013-01-04 12:00:00 a 0.749185
b -0.675521
2013-01-05 00:00:00 a 0.440266
b 0.688972
2013-01-05 12:00:00 a -0.276646
b 1.924533
[20 rows x 1 columns]
In [24]: dft2.loc['2013-01-05']
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[24]:
A
2013-01-05 00:00:00 a 0.440266
b 0.688972
2013-01-05 12:00:00 a -0.276646
b 1.924533
On other levels
In [25]: idx = pd.IndexSlice
In [26]: dft2 = dft2.swaplevel(0, 1).sort_index()
In [27]: dft2
Out[27]:
A
a 2013-01-01 00:00:00 0.156998
2013-01-01 12:00:00 1.057633
2013-01-02 00:00:00 -0.524627
2013-01-02 12:00:00 1.910759
2013-01-03 00:00:00 0.513082
2013-01-03 12:00:00 1.043945
2013-01-04 00:00:00 1.459927
... ...
b 2013-01-02 12:00:00 0.787965
2013-01-03 00:00:00 -0.546416
2013-01-03 12:00:00 2.107785
2013-01-04 00:00:00 1.015405
2013-01-04 12:00:00 -0.675521
2013-01-05 00:00:00 0.688972
2013-01-05 12:00:00 1.924533
[20 rows x 1 columns]
In [28]: dft2.loc[idx[:, '2013-01-05'], :]
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[28]:
A
a 2013-01-05 00:00:00 0.440266
2013-01-05 12:00:00 -0.276646
b 2013-01-05 00:00:00 0.688972
2013-01-05 12:00:00 1.924533
Assembling Datetimes¶
pd.to_datetime() has gained the ability to assemble datetimes from a passed in DataFrame or a dict. (GH8158).
In [29]: df = pd.DataFrame({'year': [2015, 2016],
....: 'month': [2, 3],
....: 'day': [4, 5],
....: 'hour': [2, 3]})
....:
In [30]: df
Out[30]:
day hour month year
0 4 2 2 2015
1 5 3 3 2016
Assembling using the passed frame.
In [31]: pd.to_datetime(df)
Out[31]:
0 2015-02-04 02:00:00
1 2016-03-05 03:00:00
dtype: datetime64[ns]
You can pass only the columns that you need to assemble.
In [32]: pd.to_datetime(df[['year', 'month', 'day']])
Out[32]:
0 2015-02-04
1 2016-03-05
dtype: datetime64[ns]
Other Enhancements¶
pd.read_csv()now supportsdelim_whitespace=Truefor the Python engine (GH12958)pd.read_csv()now supports opening ZIP files that contains a single CSV, via extension inference or explictcompression='zip'(GH12175)pd.read_csv()now supports opening files using xz compression, via extension inference or explicitcompression='xz'is specified;xzcompressions is also supported byDataFrame.to_csvin the same way (GH11852)pd.read_msgpack()now always gives writeable ndarrays even when compression is used (GH12359).pd.read_msgpack()now supports serializing and de-serializing categoricals with msgpack (GH12573).to_json()now supportsNDFramesthat contain categorical and sparse data (GH10778)interpolate()now supportsmethod='akima'(GH7588).pd.read_excel()now accepts path objects (e.g.pathlib.Path,py.path.local) for the file path, in line with otherread_*functions (GH12655)Added
.weekday_nameproperty as a component toDatetimeIndexand the.dtaccessor. (GH11128)Index.takenow handlesallow_fillandfill_valueconsistently (GH12631)In [33]: idx = pd.Index([1., 2., 3., 4.], dtype='float') # default, allow_fill=True, fill_value=None In [34]: idx.take([2, -1]) Out[34]: Float64Index([3.0, 4.0], dtype='float64') In [35]: idx.take([2, -1], fill_value=True) ���������������������������������������������������Out[35]: Float64Index([3.0, nan], dtype='float64')
Indexnow supports.str.get_dummies()which returnsMultiIndex, see Creating Indicator Variables (GH10008, GH10103)In [36]: idx = pd.Index(['a|b', 'a|c', 'b|c']) In [37]: idx.str.get_dummies('|') Out[37]: MultiIndex(levels=[[0, 1], [0, 1], [0, 1]], labels=[[1, 1, 0], [1, 0, 1], [0, 1, 1]], names=['a', 'b', 'c'])
pd.crosstab()has gained anormalizeargument for normalizing frequency tables (GH12569). Examples in the updated docs here..resample(..).interpolate()is now supported (GH12925).isin()now accepts passedsets(GH12988)
Sparse changes¶
These changes conform sparse handling to return the correct types and work to make a smoother experience with indexing.
SparseArray.take now returns a scalar for scalar input, SparseArray for others. Furthermore, it handles a negative indexer with the same rule as Index (GH10560, GH12796)
In [38]: s = pd.SparseArray([np.nan, np.nan, 1, 2, 3, np.nan, 4, 5, np.nan, 6])
In [39]: s.take(0)
Out[39]: nan
In [40]: s.take([1, 2, 3])
�������������Out[40]:
[nan, 1.0, 2.0]
Fill: nan
IntIndex
Indices: array([1, 2], dtype=int32)
- Bug in
SparseSeries[]indexing withEllipsisraisesKeyError(GH9467) - Bug in
SparseArray[]indexing with tuples are not handled properly (GH12966) - Bug in
SparseSeries.loc[]with list-like input raisesTypeError(GH10560) - Bug in
SparseSeries.iloc[]with scalar input may raiseIndexError(GH10560) - Bug in
SparseSeries.loc[],.iloc[]withslicereturnsSparseArray, rather thanSparseSeries(GH10560) - Bug in
SparseDataFrame.loc[],.iloc[]may results in denseSeries, rather thanSparseSeries(GH12787) - Bug in
SparseArrayaddition ignoresfill_valueof right hand side (GH12910) - Bug in
SparseArraymod raisesAttributeError(GH12910) - Bug in
SparseArraypow calculates1 ** np.nanasnp.nanwhich must be 1 (GH12910) - Bug in
SparseArraycomparison output may incorrect result or raiseValueError(GH12971) - Bug in
SparseSeries.__repr__raisesTypeErrorwhen it is longer thanmax_rows(GH10560) - Bug in
SparseSeries.shapeignoresfill_value(GH10452) - Bug in
SparseSeriesandSparseArraymay have differentdtypefrom its dense values (GH12908) - Bug in
SparseSeries.reindexincorrectly handlefill_value(GH12797) - Bug in
SparseArray.to_frame()results inDataFrame, rather thanSparseDataFrame(GH9850) - Bug in
SparseSeries.value_counts()does not countfill_value(GH6749) - Bug in
SparseArray.to_dense()does not preservedtype(GH10648) - Bug in
SparseArray.to_dense()incorrectly handlefill_value(GH12797) - Bug in
pd.concat()ofSparseSeriesresults in dense (GH10536) - Bug in
pd.concat()ofSparseDataFrameincorrectly handlefill_value(GH9765) - Bug in
pd.concat()ofSparseDataFramemay raiseAttributeError(GH12174) - Bug in
SparseArray.shift()may raiseNameErrororTypeError(GH12908)
API changes¶
.groupby(..).nth() changes¶
The index in .groupby(..).nth() output is now more consistent when the as_index argument is passed (GH11039):
In [41]: df = DataFrame({'A' : ['a', 'b', 'a'],
....: 'B' : [1, 2, 3]})
....:
In [42]: df
Out[42]:
A B
0 a 1
1 b 2
2 a 3
Previous Behavior:
In [3]: df.groupby('A', as_index=True)['B'].nth(0)
Out[3]:
0 1
1 2
Name: B, dtype: int64
In [4]: df.groupby('A', as_index=False)['B'].nth(0)
Out[4]:
0 1
1 2
Name: B, dtype: int64
New Behavior:
In [43]: df.groupby('A', as_index=True)['B'].nth(0)
Out[43]:
A
a 1
b 2
Name: B, dtype: int64
In [44]: df.groupby('A', as_index=False)['B'].nth(0)
������������������������������������������������Out[44]:
0 1
1 2
Name: B, dtype: int64
Furthermore, previously, a .groupby would always sort, regardless if sort=False was passed with .nth().
In [45]: np.random.seed(1234)
In [46]: df = pd.DataFrame(np.random.randn(100, 2), columns=['a', 'b'])
In [47]: df['c'] = np.random.randint(0, 4, 100)
Previous Behavior:
In [4]: df.groupby('c', sort=True).nth(1)
Out[4]:
a b
c
0 -0.334077 0.002118
1 0.036142 -2.074978
2 -0.720589 0.887163
3 0.859588 -0.636524
In [5]: df.groupby('c', sort=False).nth(1)
Out[5]:
a b
c
0 -0.334077 0.002118
1 0.036142 -2.074978
2 -0.720589 0.887163
3 0.859588 -0.636524
New Behavior:
In [48]: df.groupby('c', sort=True).nth(1)
Out[48]:
a b
c
0 -0.334077 0.002118
1 0.036142 -2.074978
2 -0.720589 0.887163
3 0.859588 -0.636524
In [49]: df.groupby('c', sort=False).nth(1)
����������������������������������������������������������������������������������������������������������������������������������������������Out[49]:
a b
c
2 -0.720589 0.887163
3 0.859588 -0.636524
0 -0.334077 0.002118
1 0.036142 -2.074978
numpy function compatibility¶
Compatibility between pandas array-like methods (e.g. sum and take) and their numpy
counterparts has been greatly increased by augmenting the signatures of the pandas methods so
as to accept arguments that can be passed in from numpy, even if they are not necessarily
used in the pandas implementation (GH12644, GH12638, GH12687)
.searchsorted()forIndexandTimedeltaIndexnow accept asorterargument to maintain compatibility with numpy’ssearchsortedfunction (GH12238)- Bug in numpy compatibility of
np.round()on aSeries(GH12600)
An example of this signature augmentation is illustrated below:
In [50]: sp = pd.SparseDataFrame([1, 2, 3])
In [51]: sp
Out[51]:
0
0 1
1 2
2 3
Previous behaviour:
In [2]: np.cumsum(sp, axis=0)
...
TypeError: cumsum() takes at most 2 arguments (4 given)
New behaviour:
In [52]: np.cumsum(sp, axis=0)
Out[52]:
0
0 1
1 3
2 6
Using .apply on groupby resampling¶
Using apply on resampling groupby operations (using a pd.TimeGrouper) now has the same output types as similar apply calls on other groupby operations. (GH11742).
In [53]: df = pd.DataFrame({'date': pd.to_datetime(['10/10/2000', '11/10/2000']),
....: 'value': [10, 13]})
....:
In [54]: df
Out[54]:
date value
0 2000-10-10 10
1 2000-11-10 13
Previous behavior:
In [1]: df.groupby(pd.TimeGrouper(key='date', freq='M')).apply(lambda x: x.value.sum())
Out[1]:
...
TypeError: cannot concatenate a non-NDFrame object
# Output is a Series
In [2]: df.groupby(pd.TimeGrouper(key='date', freq='M')).apply(lambda x: x[['value']].sum())
Out[2]:
date
2000-10-31 value 10
2000-11-30 value 13
dtype: int64
New Behavior:
# Output is a Series
In [55]: df.groupby(pd.TimeGrouper(key='date', freq='M')).apply(lambda x: x.value.sum())
Out[55]:
date
2000-10-31 10
2000-11-30 13
Freq: M, dtype: int64
# Output is a DataFrame
In [56]: df.groupby(pd.TimeGrouper(key='date', freq='M')).apply(lambda x: x[['value']].sum())
�����������������������������������������������������������������������Out[56]:
value
date
2000-10-31 10
2000-11-30 13
Changes in read_csv exceptions¶
In order to standardize the read_csv API for both the c and python engines, both will now raise an
EmptyDataError, a subclass of ValueError, in response to empty columns or header (GH12493, GH12506)
Previous behaviour:
In [1]: df = pd.read_csv(StringIO(''), engine='c')
...
ValueError: No columns to parse from file
In [2]: df = pd.read_csv(StringIO(''), engine='python')
...
StopIteration
New behaviour:
In [1]: df = pd.read_csv(StringIO(''), engine='c')
...
pandas.io.common.EmptyDataError: No columns to parse from file
In [2]: df = pd.read_csv(StringIO(''), engine='python')
...
pandas.io.common.EmptyDataError: No columns to parse from file
In addition to this error change, several others have been made as well:
CParserErrornow sub-classesValueErrorinstead of just aException(GH12551)- A
CParserErroris now raised instead of a genericExceptioninread_csvwhen thecengine cannot parse a column (GH12506) - A
ValueErroris now raised instead of a genericExceptioninread_csvwhen thecengine encounters aNaNvalue in an integer column (GH12506) - A
ValueErroris now raised instead of a genericExceptioninread_csvwhentrue_valuesis specified, and thecengine encounters an element in a column containing unencodable bytes (GH12506) pandas.parser.OverflowErrorexception has been removed and has been replaced with Python’s built-inOverflowErrorexception (GH12506)pd.read_csv()no longer allows a combination of strings and integers for theusecolsparameter (GH12678)
to_datetime error changes¶
Bugs in pd.to_datetime() when passing a unit with convertible entries and errors='coerce' or non-convertible with errors='ignore'. Furthermore, an OutOfBoundsDateime exception will be raised when an out-of-range value is encountered for that unit when errors='raise'. (GH11758, GH13052, GH13059)
Previous behaviour:
In [27]: pd.to_datetime(1420043460, unit='s', errors='coerce')
Out[27]: NaT
In [28]: pd.to_datetime(11111111, unit='D', errors='ignore')
OverflowError: Python int too large to convert to C long
In [29]: pd.to_datetime(11111111, unit='D', errors='raise')
OverflowError: Python int too large to convert to C long
New behaviour:
In [2]: pd.to_datetime(1420043460, unit='s', errors='coerce')
Out[2]: Timestamp('2014-12-31 16:31:00')
In [3]: pd.to_datetime(11111111, unit='D', errors='ignore')
Out[3]: 11111111
In [4]: pd.to_datetime(11111111, unit='D', errors='raise')
OutOfBoundsDatetime: cannot convert input with unit 'D'
Other API changes¶
.swaplevel()forSeries,DataFrame,Panel, andMultiIndexnow features defaults for its first two parametersiandjthat swap the two innermost levels of the index. (GH12934).searchsorted()forIndexandTimedeltaIndexnow accept asorterargument to maintain compatibility with numpy’ssearchsortedfunction (GH12238)PeriodandPeriodIndexnow raisesIncompatibleFrequencyerror which inheritsValueErrorrather than rawValueError(GH12615)Series.applyfor category dtype now applies the passed function to each of the.categories(and not the.codes), and returns acategorydtype if possible (GH12473)read_csvwill now raise aTypeErrorifparse_datesis neither a boolean, list, or dictionary (matches the doc-string) (GH5636)- The default for
.query()/.eval()is nowengine=None, which will usenumexprif it’s installed; otherwise it will fallback to thepythonengine. This mimics the pre-0.18.1 behavior ifnumexpris installed (and which, previously, if numexpr was not installed,.query()/.eval()would raise). (GH12749) pd.show_versions()now includespandas_datareaderversion (GH12740)- Provide a proper
__name__and__qualname__attributes for generic functions (GH12021) pd.concat(ignore_index=True)now usesRangeIndexas default (GH12695)pd.merge()andDataFrame.join()will show aUserWarningwhen merging/joining a single- with a multi-leveled dataframe (GH9455, GH12219)- Compat with
scipy> 0.17 for deprecatedpiecewise_polynomialinterpolation method; support for the replacementfrom_derivativesmethod (GH12887)
Performance Improvements¶
- Improved speed of SAS reader (GH12656, GH12961)
- Performance improvements in
.groupby(..).cumcount()(GH11039) - Improved memory usage in
pd.read_csv()when usingskiprows=an_integer(GH13005) - Improved performance of
DataFrame.to_sqlwhen checking case sensitivity for tables. Now only checks if table has been created correctly when table name is not lower case. (GH12876) - Improved performance of
Periodconstruction and time series plotting (GH12903, GH11831). - Improved performance of
.str.encode()and.str.decode()methods (GH13008) - Improved performance of
to_numericif input is numeric dtype (GH12777) - Improved performance of sparse arithmetic with
IntIndex(GH13036)
Bug Fixes¶
usecolsparameter inpd.read_csvis now respected even when the lines of a CSV file are not even (GH12203)- Bug in
groupby.transform(..)whenaxis=1is specified with a non-monotonic ordered index (GH12713) - Bug in
PeriodandPeriodIndexcreation raisesKeyErroriffreq="Minute"is specified. Note that “Minute” freq is deprecated in v0.17.0, and recommended to usefreq="T"instead (GH11854) - Bug in
.resample(...).count()with aPeriodIndexalways raising aTypeError(GH12774) - Bug in
.resample(...)with aPeriodIndexcasting to aDatetimeIndexwhen empty (GH12868) - Bug in
.resample(...)with aPeriodIndexwhen resampling to an existing frequency (GH12770) - Bug in printing data which contains
Periodwith differentfreqraisesValueError(GH12615) - Bug in
Seriesconstruction withCategoricalanddtype='category'is specified (GH12574) - Bugs in concatenation with a coercable dtype was too aggressive, resulting in different dtypes in outputformatting when an object was longer than
display.max_rows(GH12411, GH12045, GH11594, GH10571, GH12211) - Bug in
float_formatoption with option not being validated as a callable. (GH12706) - Bug in
GroupBy.filterwhendropna=Falseand no groups fulfilled the criteria (GH12768) - Bug in
__name__of.cum*functions (GH12021) - Bug in
.astype()of aFloat64Inde/Int64Indexto anInt64Index(GH12881) - Bug in roundtripping an integer based index in
.to_json()/.read_json()whenorient='index'(the default) (GH12866) - Bug in plotting
Categoricaldtypes cause error when attempting stacked bar plot (GH13019) - Compat with >=
numpy1.11 forNaTcomparions (GH12969) - Bug in
.drop()with a non-uniqueMultiIndex. (GH12701) - Bug in
.concatof datetime tz-aware and naive DataFrames (GH12467) - Bug in correctly raising a
ValueErrorin.resample(..).fillna(..)when passing a non-string (GH12952) - Bug fixes in various encoding and header processing issues in
pd.read_sas()(GH12659, GH12654, GH12647, GH12809) - Bug in
pd.crosstab()where would silently ignoreaggfuncifvalues=None(GH12569). - Potential segfault in
DataFrame.to_jsonwhen serialisingdatetime.time(GH11473). - Potential segfault in
DataFrame.to_jsonwhen attempting to serialise 0d array (GH11299). - Segfault in
to_jsonwhen attempting to serialise aDataFrameorSerieswith non-ndarray values; now supports serialization ofcategory,sparse, anddatetime64[ns, tz]dtypes (GH10778). - Bug in
DataFrame.to_jsonwith unsupported dtype not passed to default handler (GH12554). - Bug in
.alignnot returning the sub-class (GH12983) - Bug in aligning a
Serieswith aDataFrame(GH13037) - Bug in
ABCPanelin whichPanel4Dwas not being considered as a valid instance of this generic type (GH12810) - Bug in consistency of
.nameon.groupby(..).apply(..)cases (GH12363) - Bug in
Timestamp.__repr__that causedpprintto fail in nested structures (GH12622) - Bug in
Timedelta.minandTimedelta.max, the properties now report the true minimum/maximumtimedeltasas recognized by pandas. See the documentation. (GH12727) - Bug in
.quantile()with interpolation may coerce tofloatunexpectedly (GH12772) - Bug in
.quantile()with emptySeriesmay return scalar rather than emptySeries(GH12772) - Bug in
.locwith out-of-bounds in a large indexer would raiseIndexErrorrather thanKeyError(GH12527) - Bug in resampling when using a
TimedeltaIndexand.asfreq(), would previously not include the final fencepost (GH12926) - Bug in equality testing with a
Categoricalin aDataFrame(GH12564) - Bug in
GroupBy.first(),.last()returns incorrect row whenTimeGrouperis used (GH7453) - Bug in
pd.read_csv()with thecengine when specifyingskiprowswith newlines in quoted items (GH10911, GH12775) - Bug in
DataFrametimezone lost when assigning tz-aware datetimeSerieswith alignment (GH12981) - Bug in
.value_counts()whennormalize=Trueanddropna=Truewhere nulls still contributed to the normalized count (GH12558) - Bug in
Series.value_counts()loses name if its dtype iscategory(GH12835) - Bug in
Series.value_counts()loses timezone info (GH12835) - Bug in
Series.value_counts(normalize=True)withCategoricalraisesUnboundLocalError(GH12835) - Bug in
Panel.fillna()ignoringinplace=True(GH12633) - Bug in
pd.read_csv()when specifyingnames,usecols, andparse_datessimultaneously with thecengine (GH9755) - Bug in
pd.read_csv()when specifyingdelim_whitespace=Trueandlineterminatorsimultaneously with thecengine (GH12912) - Bug in
Series.rename,DataFrame.renameandDataFrame.rename_axisnot treatingSeriesas mappings to relabel (GH12623). - Clean in
.rolling.minand.rolling.maxto enhance dtype handling (GH12373) - Bug in
groupbywhere complex types are coerced to float (GH12902) - Bug in
Series.mapraisesTypeErrorif its dtype iscategoryor tz-awaredatetime(GH12473) - Bugs on 32bit platforms for some test comparisons (GH12972)
- Bug in index coercion when falling back from
RangeIndexconstruction (GH12893) - Better error message in window functions when invalid argument (e.g. a float window) is passed (GH12669)
- Bug in slicing subclassed
DataFramedefined to return subclassedSeriesmay return normalSeries(GH11559) - Bug in
.straccessor methods may raiseValueErrorif input hasnameand the result isDataFrameorMultiIndex(GH12617) - Bug in
DataFrame.last_valid_index()andDataFrame.first_valid_index()on empty frames (GH12800) - Bug in
CategoricalIndex.get_locreturns different result from regularIndex(GH12531) - Bug in
PeriodIndex.resamplewhere name not propagated (GH12769) - Bug in
date_rangeclosedkeyword and timezones (GH12684). - Bug in
pd.concatraisesAttributeErrorwhen input data contains tz-aware datetime and timedelta (GH12620) - Bug in
pd.concatdid not handle emptySeriesproperly (GH11082) - Bug in
.plot.baralginment whenwidthis specified withint(GH12979) - Bug in
fill_valueis ignored if the argument to a binary operator is a constant (GH12723) - Bug in
pd.read_html()when using bs4 flavor and parsing table with a header and only one column (GH9178) - Bug in
.pivot_tablewhenmargins=Trueanddropna=Truewhere nulls still contributed to margin count (GH12577) - Bug in
.pivot_tablewhendropna=Falsewhere table index/column names disappear (GH12133) - Bug in
pd.crosstab()whenmargins=Trueanddropna=Falsewhich raised (GH12642) - Bug in
Series.namewhennameattribute can be a hashable type (GH12610) - Bug in
.describe()resets categorical columns information (GH11558) - Bug where
loffsetargument was not applied when callingresample().count()on a timeseries (GH12725) pd.read_excel()now accepts column names associated with keyword argumentnames(GH12870)- Bug in
pd.to_numeric()withIndexreturnsnp.ndarray, rather thanIndex(GH12777) - Bug in
pd.to_numeric()with datetime-like may raiseTypeError(GH12777) - Bug in
pd.to_numeric()with scalar raisesValueError(GH12777)
v0.18.0 (March 13, 2016)¶
This is a major release from 0.17.1 and includes a small number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
Warning
pandas >= 0.18.0 no longer supports compatibility with Python version 2.6 and 3.3 (GH7718, GH11273)
Warning
numexpr version 2.4.4 will now show a warning and not be used as a computation back-end for pandas because of some buggy behavior. This does not affect other versions (>= 2.1 and >= 2.4.6). (GH12489)
Highlights include:
- Moving and expanding window functions are now methods on Series and DataFrame,
similar to
.groupby, see here. - Adding support for a
RangeIndexas a specialized form of theInt64Indexfor memory savings, see here. - API breaking change to the
.resamplemethod to make it more.groupbylike, see here. - Removal of support for positional indexing with floats, which was deprecated
since 0.14.0. This will now raise a
TypeError, see here. - The
.to_xarray()function has been added for compatibility with the xarray package, see here. - The
read_sasfunction has been enhanced to readsas7bdatfiles, see here. - Addition of the .str.extractall() method, and API changes to the .str.extract() method and .str.cat() method.
pd.test()top-level nose test runner is available (GH4327).
Check the API Changes and deprecations before updating.
What’s new in v0.18.0
- New features
- Window functions are now methods
- Changes to rename
- Range Index
- Changes to str.extract
- Addition of str.extractall
- Changes to str.cat
- Datetimelike rounding
- Formatting of Integers in FloatIndex
- Changes to dtype assignment behaviors
- to_xarray
- Latex Representation
pd.read_sas()changes- Other enhancements
- Backwards incompatible API changes
- Performance Improvements
- Bug Fixes
New features¶
Window functions are now methods¶
Window functions have been refactored to be methods on Series/DataFrame objects, rather than top-level functions, which are now deprecated. This allows these window-type functions, to have a similar API to that of .groupby. See the full documentation here (GH11603, GH12373)
In [1]: np.random.seed(1234)
In [2]: df = pd.DataFrame({'A' : range(10), 'B' : np.random.randn(10)})
In [3]: df
Out[3]:
A B
0 0 0.471435
1 1 -1.190976
2 2 1.432707
3 3 -0.312652
4 4 -0.720589
5 5 0.887163
6 6 0.859588
7 7 -0.636524
8 8 0.015696
9 9 -2.242685
Previous Behavior:
In [8]: pd.rolling_mean(df,window=3)
FutureWarning: pd.rolling_mean is deprecated for DataFrame and will be removed in a future version, replace with
DataFrame.rolling(window=3,center=False).mean()
Out[8]:
A B
0 NaN NaN
1 NaN NaN
2 1 0.237722
3 2 -0.023640
4 3 0.133155
5 4 -0.048693
6 5 0.342054
7 6 0.370076
8 7 0.079587
9 8 -0.954504
New Behavior:
In [4]: r = df.rolling(window=3)
These show a descriptive repr
In [5]: r
Out[5]: Rolling [window=3,center=False,axis=0]
with tab-completion of available methods and properties.
In [9]: r.
r.A r.agg r.apply r.count r.exclusions r.max r.median r.name r.skew r.sum
r.B r.aggregate r.corr r.cov r.kurt r.mean r.min r.quantile r.std r.var
The methods operate on the Rolling object itself
In [6]: r.mean()
Out[6]:
A B
0 NaN NaN
1 NaN NaN
2 1.0 0.237722
3 2.0 -0.023640
4 3.0 0.133155
5 4.0 -0.048693
6 5.0 0.342054
7 6.0 0.370076
8 7.0 0.079587
9 8.0 -0.954504
They provide getitem accessors
In [7]: r['A'].mean()
Out[7]:
0 NaN
1 NaN
2 1.0
3 2.0
4 3.0
5 4.0
6 5.0
7 6.0
8 7.0
9 8.0
Name: A, dtype: float64
And multiple aggregations
In [8]: r.agg({'A' : ['mean','std'],
...: 'B' : ['mean','std']})
...:
Out[8]:
A B
mean std mean std
0 NaN NaN NaN NaN
1 NaN NaN NaN NaN
2 1.0 1.0 0.237722 1.327364
3 2.0 1.0 -0.023640 1.335505
4 3.0 1.0 0.133155 1.143778
5 4.0 1.0 -0.048693 0.835747
6 5.0 1.0 0.342054 0.920379
7 6.0 1.0 0.370076 0.871850
8 7.0 1.0 0.079587 0.750099
9 8.0 1.0 -0.954504 1.162285
Changes to rename¶
Series.rename and NDFrame.rename_axis can now take a scalar or list-like
argument for altering the Series or axis name, in addition to their old behaviors of altering labels. (GH9494, GH11965)
In [9]: s = pd.Series(np.random.randn(5))
In [10]: s.rename('newname')
Out[10]:
0 1.150036
1 0.991946
2 0.953324
3 -2.021255
4 -0.334077
Name: newname, dtype: float64
In [11]: df = pd.DataFrame(np.random.randn(5, 2))
In [12]: (df.rename_axis("indexname")
....: .rename_axis("columns_name", axis="columns"))
....:
Out[12]:
columns_name 0 1
indexname
0 0.002118 0.405453
1 0.289092 1.321158
2 -1.546906 -0.202646
3 -0.655969 0.193421
4 0.553439 1.318152
The new functionality works well in method chains. Previously these methods only accepted functions or dicts mapping a label to a new label. This continues to work as before for function or dict-like values.
Range Index¶
A RangeIndex has been added to the Int64Index sub-classes to support a memory saving alternative for common use cases. This has a similar implementation to the python range object (xrange in python 2), in that it only stores the start, stop, and step values for the index. It will transparently interact with the user API, converting to Int64Index if needed.
This will now be the default constructed index for NDFrame objects, rather than previous an Int64Index. (GH939, GH12070, GH12071, GH12109, GH12888)
Previous Behavior:
In [3]: s = pd.Series(range(1000))
In [4]: s.index
Out[4]:
Int64Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9,
...
990, 991, 992, 993, 994, 995, 996, 997, 998, 999], dtype='int64', length=1000)
In [6]: s.index.nbytes
Out[6]: 8000
New Behavior:
In [13]: s = pd.Series(range(1000))
In [14]: s.index
Out[14]: RangeIndex(start=0, stop=1000, step=1)
In [15]: s.index.nbytes
������������������������������������������������Out[15]: 80
Changes to str.extract¶
The .str.extract method takes a regular expression with capture groups, finds the first match in each subject string, and returns the contents of the capture groups (GH11386).
In v0.18.0, the expand argument was added to
extract.
expand=False: it returns aSeries,Index, orDataFrame, depending on the subject and regular expression pattern (same behavior as pre-0.18.0).expand=True: it always returns aDataFrame, which is more consistent and less confusing from the perspective of a user.
Currently the default is expand=None which gives a FutureWarning and uses expand=False. To avoid this warning, please explicitly specify expand.
In [1]: pd.Series(['a1', 'b2', 'c3']).str.extract('[ab](\d)', expand=None)
FutureWarning: currently extract(expand=None) means expand=False (return Index/Series/DataFrame)
but in a future version of pandas this will be changed to expand=True (return DataFrame)
Out[1]:
0 1
1 2
2 NaN
dtype: object
Extracting a regular expression with one group returns a Series if
expand=False.
In [16]: pd.Series(['a1', 'b2', 'c3']).str.extract('[ab](\d)', expand=False)
Out[16]:
0 1
1 2
2 NaN
dtype: object
It returns a DataFrame with one column if expand=True.
In [17]: pd.Series(['a1', 'b2', 'c3']).str.extract('[ab](\d)', expand=True)
Out[17]:
0
0 1
1 2
2 NaN
Calling on an Index with a regex with exactly one capture group
returns an Index if expand=False.
In [18]: s = pd.Series(["a1", "b2", "c3"], ["A11", "B22", "C33"])
In [19]: s.index
Out[19]: Index(['A11', 'B22', 'C33'], dtype='object')
In [20]: s.index.str.extract("(?P<letter>[a-zA-Z])", expand=False)
������������������������������������������������������Out[20]: Index(['A', 'B', 'C'], dtype='object', name='letter')
It returns a DataFrame with one column if expand=True.
In [21]: s.index.str.extract("(?P<letter>[a-zA-Z])", expand=True)
Out[21]:
letter
0 A
1 B
2 C
Calling on an Index with a regex with more than one capture group
raises ValueError if expand=False.
>>> s.index.str.extract("(?P<letter>[a-zA-Z])([0-9]+)", expand=False)
ValueError: only one regex group is supported with Index
It returns a DataFrame if expand=True.
In [22]: s.index.str.extract("(?P<letter>[a-zA-Z])([0-9]+)", expand=True)
Out[22]:
letter 1
0 A 11
1 B 22
2 C 33
In summary, extract(expand=True) always returns a DataFrame
with a row for every subject string, and a column for every capture
group.
Addition of str.extractall¶
The .str.extractall method was added
(GH11386). Unlike extract, which returns only the first
match.
In [23]: s = pd.Series(["a1a2", "b1", "c1"], ["A", "B", "C"])
In [24]: s
Out[24]:
A a1a2
B b1
C c1
dtype: object
In [25]: s.str.extract("(?P<letter>[ab])(?P<digit>\d)", expand=False)
������������������������������������������������������Out[25]:
letter digit
A a 1
B b 1
C NaN NaN
The extractall method returns all matches.
In [26]: s.str.extractall("(?P<letter>[ab])(?P<digit>\d)")
Out[26]:
letter digit
match
A 0 a 1
1 a 2
B 0 b 1
Changes to str.cat¶
The method .str.cat() concatenates the members of a Series. Before, if NaN values were present in the Series, calling .str.cat() on it would return NaN, unlike the rest of the Series.str.* API. This behavior has been amended to ignore NaN values by default. (GH11435).
A new, friendlier ValueError is added to protect against the mistake of supplying the sep as an arg, rather than as a kwarg. (GH11334).
In [27]: pd.Series(['a','b',np.nan,'c']).str.cat(sep=' ')
Out[27]: 'a b c'
In [28]: pd.Series(['a','b',np.nan,'c']).str.cat(sep=' ', na_rep='?')
�����������������Out[28]: 'a b ? c'
In [2]: pd.Series(['a','b',np.nan,'c']).str.cat(' ')
ValueError: Did you mean to supply a `sep` keyword?
Datetimelike rounding¶
DatetimeIndex, Timestamp, TimedeltaIndex, Timedelta have gained the .round(), .floor() and .ceil() method for datetimelike rounding, flooring and ceiling. (GH4314, GH11963)
Naive datetimes
In [29]: dr = pd.date_range('20130101 09:12:56.1234', periods=3)
In [30]: dr
Out[30]:
DatetimeIndex(['2013-01-01 09:12:56.123400', '2013-01-02 09:12:56.123400',
'2013-01-03 09:12:56.123400'],
dtype='datetime64[ns]', freq='D')
In [31]: dr.round('s')
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[31]:
DatetimeIndex(['2013-01-01 09:12:56', '2013-01-02 09:12:56',
'2013-01-03 09:12:56'],
dtype='datetime64[ns]', freq=None)
# Timestamp scalar
In [32]: dr[0]
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[32]: Timestamp('2013-01-01 09:12:56.123400', freq='D')
In [33]: dr[0].round('10s')
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[33]: Timestamp('2013-01-01 09:13:00')
Tz-aware are rounded, floored and ceiled in local times
In [34]: dr = dr.tz_localize('US/Eastern')
In [35]: dr
Out[35]:
DatetimeIndex(['2013-01-01 09:12:56.123400-05:00',
'2013-01-02 09:12:56.123400-05:00',
'2013-01-03 09:12:56.123400-05:00'],
dtype='datetime64[ns, US/Eastern]', freq='D')
In [36]: dr.round('s')
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[36]:
DatetimeIndex(['2013-01-01 09:12:56-05:00', '2013-01-02 09:12:56-05:00',
'2013-01-03 09:12:56-05:00'],
dtype='datetime64[ns, US/Eastern]', freq=None)
Timedeltas
In [37]: t = timedelta_range('1 days 2 hr 13 min 45 us',periods=3,freq='d')
In [38]: t
Out[38]:
TimedeltaIndex(['1 days 02:13:00.000045', '2 days 02:13:00.000045',
'3 days 02:13:00.000045'],
dtype='timedelta64[ns]', freq='D')
In [39]: t.round('10min')
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[39]: TimedeltaIndex(['1 days 02:10:00', '2 days 02:10:00', '3 days 02:10:00'], dtype='timedelta64[ns]', freq=None)
# Timedelta scalar
In [40]: t[0]
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[40]: Timedelta('1 days 02:13:00.000045')
In [41]: t[0].round('2h')
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[41]: Timedelta('1 days 02:00:00')
In addition, .round(), .floor() and .ceil() will be available thru the .dt accessor of Series.
In [42]: s = pd.Series(dr)
In [43]: s
Out[43]:
0 2013-01-01 09:12:56.123400-05:00
1 2013-01-02 09:12:56.123400-05:00
2 2013-01-03 09:12:56.123400-05:00
dtype: datetime64[ns, US/Eastern]
In [44]: s.dt.round('D')
�����������������������������������������������������������������������������������������������������������������������������������������������������������Out[44]:
0 2013-01-01 00:00:00-05:00
1 2013-01-02 00:00:00-05:00
2 2013-01-03 00:00:00-05:00
dtype: datetime64[ns, US/Eastern]
Formatting of Integers in FloatIndex¶
Integers in FloatIndex, e.g. 1., are now formatted with a decimal point and a 0 digit, e.g. 1.0 (GH11713)
This change not only affects the display to the console, but also the output of IO methods like .to_csv or .to_html.
Previous Behavior:
In [2]: s = pd.Series([1,2,3], index=np.arange(3.))
In [3]: s
Out[3]:
0 1
1 2
2 3
dtype: int64
In [4]: s.index
Out[4]: Float64Index([0.0, 1.0, 2.0], dtype='float64')
In [5]: print(s.to_csv(path=None))
0,1
1,2
2,3
New Behavior:
In [45]: s = pd.Series([1,2,3], index=np.arange(3.))
In [46]: s
Out[46]:
0.0 1
1.0 2
2.0 3
dtype: int64
In [47]: s.index
��������������������������������������������������Out[47]: Float64Index([0.0, 1.0, 2.0], dtype='float64')
In [48]: print(s.to_csv(path=None))
����������������������������������������������������������������������������������������������������������0.0,1
1.0,2
2.0,3
Changes to dtype assignment behaviors¶
When a DataFrame’s slice is updated with a new slice of the same dtype, the dtype of the DataFrame will now remain the same. (GH10503)
Previous Behavior:
In [5]: df = pd.DataFrame({'a': [0, 1, 1],
'b': pd.Series([100, 200, 300], dtype='uint32')})
In [7]: df.dtypes
Out[7]:
a int64
b uint32
dtype: object
In [8]: ix = df['a'] == 1
In [9]: df.loc[ix, 'b'] = df.loc[ix, 'b']
In [11]: df.dtypes
Out[11]:
a int64
b int64
dtype: object
New Behavior:
In [49]: df = pd.DataFrame({'a': [0, 1, 1],
....: 'b': pd.Series([100, 200, 300], dtype='uint32')})
....:
In [50]: df.dtypes
Out[50]:
a int64
b uint32
dtype: object
In [51]: ix = df['a'] == 1
In [52]: df.loc[ix, 'b'] = df.loc[ix, 'b']
In [53]: df.dtypes
Out[53]:
a int64
b uint32
dtype: object
When a DataFrame’s integer slice is partially updated with a new slice of floats that could potentially be downcasted to integer without losing precision, the dtype of the slice will be set to float instead of integer.
Previous Behavior:
In [4]: df = pd.DataFrame(np.array(range(1,10)).reshape(3,3),
columns=list('abc'),
index=[[4,4,8], [8,10,12]])
In [5]: df
Out[5]:
a b c
4 8 1 2 3
10 4 5 6
8 12 7 8 9
In [7]: df.ix[4, 'c'] = np.array([0., 1.])
In [8]: df
Out[8]:
a b c
4 8 1 2 0
10 4 5 1
8 12 7 8 9
New Behavior:
In [54]: df = pd.DataFrame(np.array(range(1,10)).reshape(3,3),
....: columns=list('abc'),
....: index=[[4,4,8], [8,10,12]])
....:
In [55]: df
Out[55]:
a b c
4 8 1 2 3
10 4 5 6
8 12 7 8 9
In [56]: df.loc[4, 'c'] = np.array([0., 1.])
In [57]: df
Out[57]:
a b c
4 8 1 2 0.0
10 4 5 1.0
8 12 7 8 9.0
to_xarray¶
In a future version of pandas, we will be deprecating Panel and other > 2 ndim objects. In order to provide for continuity,
all NDFrame objects have gained the .to_xarray() method in order to convert to xarray objects, which has
a pandas-like interface for > 2 ndim. (GH11972)
See the xarray full-documentation here.
In [1]: p = Panel(np.arange(2*3*4).reshape(2,3,4))
In [2]: p.to_xarray()
Out[2]:
<xarray.DataArray (items: 2, major_axis: 3, minor_axis: 4)>
array([[[ 0, 1, 2, 3],
[ 4, 5, 6, 7],
[ 8, 9, 10, 11]],
[[12, 13, 14, 15],
[16, 17, 18, 19],
[20, 21, 22, 23]]])
Coordinates:
* items (items) int64 0 1
* major_axis (major_axis) int64 0 1 2
* minor_axis (minor_axis) int64 0 1 2 3
Latex Representation¶
DataFrame has gained a ._repr_latex_() method in order to allow for conversion to latex in a ipython/jupyter notebook using nbconvert. (GH11778)
Note that this must be activated by setting the option pd.display.latex.repr=True (GH12182)
For example, if you have a jupyter notebook you plan to convert to latex using nbconvert, place the statement pd.display.latex.repr=True in the first cell to have the contained DataFrame output also stored as latex.
The options display.latex.escape and display.latex.longtable have also been added to the configuration and are used automatically by the to_latex
method. See the available options docs for more info.
pd.read_sas() changes¶
read_sas has gained the ability to read SAS7BDAT files, including compressed files. The files can be read in entirety, or incrementally. For full details see here. (GH4052)
Other enhancements¶
- Handle truncated floats in SAS xport files (GH11713)
- Added option to hide index in
Series.to_string(GH11729) read_excelnow supports s3 urls of the formats3://bucketname/filename(GH11447)- add support for
AWS_S3_HOSTenv variable when reading from s3 (GH12198) - A simple version of
Panel.round()is now implemented (GH11763) - For Python 3.x,
round(DataFrame),round(Series),round(Panel)will work (GH11763) sys.getsizeof(obj)returns the memory usage of a pandas object, including the values it contains (GH11597)Seriesgained anis_uniqueattribute (GH11946)DataFrame.quantileandSeries.quantilenow acceptinterpolationkeyword (GH10174).- Added
DataFrame.style.formatfor more flexible formatting of cell values (GH11692) DataFrame.select_dtypesnow allows thenp.float16typecode (GH11990)pivot_table()now accepts most iterables for thevaluesparameter (GH12017)- Added Google
BigQueryservice account authentication support, which enables authentication on remote servers. (GH11881, GH12572). For further details see here HDFStoreis now iterable:for k in storeis equivalent tofor k in store.keys()(GH12221).- Add missing methods/fields to
.dtforPeriod(GH8848) - The entire codebase has been
PEP-ified (GH12096)
Backwards incompatible API changes¶
- the leading whitespaces have been removed from the output of
.to_string(index=False)method (GH11833) - the
outparameter has been removed from theSeries.round()method. (GH11763) DataFrame.round()leaves non-numeric columns unchanged in its return, rather than raises. (GH11885)DataFrame.head(0)andDataFrame.tail(0)return empty frames, rather thanself. (GH11937)Series.head(0)andSeries.tail(0)return empty series, rather thanself. (GH11937)to_msgpackandread_msgpackencoding now defaults to'utf-8'. (GH12170)- the order of keyword arguments to text file parsing functions (
.read_csv(),.read_table(),.read_fwf()) changed to group related arguments. (GH11555) NaTType.isoformatnow returns the string'NaTto allow the result to be passed to the constructor ofTimestamp. (GH12300)
NaT and Timedelta operations¶
NaT and Timedelta have expanded arithmetic operations, which are extended to Series
arithmetic where applicable. Operations defined for datetime64[ns] or timedelta64[ns]
are now also defined for NaT (GH11564).
NaT now supports arithmetic operations with integers and floats.
In [58]: pd.NaT * 1
Out[58]: NaT
In [59]: pd.NaT * 1.5
�������������Out[59]: NaT
In [60]: pd.NaT / 2
��������������������������Out[60]: NaT
In [61]: pd.NaT * np.nan
���������������������������������������Out[61]: NaT
NaT defines more arithmetic operations with datetime64[ns] and timedelta64[ns].
In [62]: pd.NaT / pd.NaT
Out[62]: nan
In [63]: pd.Timedelta('1s') / pd.NaT
�������������Out[63]: nan
NaT may represent either a datetime64[ns] null or a timedelta64[ns] null.
Given the ambiguity, it is treated as a timedelta64[ns], which allows more operations
to succeed.
In [64]: pd.NaT + pd.NaT
Out[64]: NaT
# same as
In [65]: pd.Timedelta('1s') + pd.Timedelta('1s')
�������������Out[65]: Timedelta('0 days 00:00:02')
as opposed to
In [3]: pd.Timestamp('19900315') + pd.Timestamp('19900315')
TypeError: unsupported operand type(s) for +: 'Timestamp' and 'Timestamp'
However, when wrapped in a Series whose dtype is datetime64[ns] or timedelta64[ns],
the dtype information is respected.
In [1]: pd.Series([pd.NaT], dtype='<M8[ns]') + pd.Series([pd.NaT], dtype='<M8[ns]')
TypeError: can only operate on a datetimes for subtraction,
but the operator [__add__] was passed
In [66]: pd.Series([pd.NaT], dtype='<m8[ns]') + pd.Series([pd.NaT], dtype='<m8[ns]')
Out[66]:
0 NaT
dtype: timedelta64[ns]
Timedelta division by floats now works.
In [67]: pd.Timedelta('1s') / 2.0
Out[67]: Timedelta('0 days 00:00:00.500000')
Subtraction by Timedelta in a Series by a Timestamp works (GH11925)
In [68]: ser = pd.Series(pd.timedelta_range('1 day', periods=3))
In [69]: ser
Out[69]:
0 1 days
1 2 days
2 3 days
dtype: timedelta64[ns]
In [70]: pd.Timestamp('2012-01-01') - ser
������������������������������������������������������������������Out[70]:
0 2011-12-31
1 2011-12-30
2 2011-12-29
dtype: datetime64[ns]
NaT.isoformat() now returns 'NaT'. This change allows allows
pd.Timestamp to rehydrate any timestamp like object from its isoformat
(GH12300).
Changes to msgpack¶
Forward incompatible changes in msgpack writing format were made over 0.17.0 and 0.18.0; older versions of pandas cannot read files packed by newer versions (GH12129, GH10527)
Bugs in to_msgpack and read_msgpack introduced in 0.17.0 and fixed in 0.18.0, caused files packed in Python 2 unreadable by Python 3 (GH12142). The following table describes the backward and forward compat of msgpacks.
Warning
| Packed with | Can be unpacked with |
|---|---|
| pre-0.17 / Python 2 | any |
| pre-0.17 / Python 3 | any |
| 0.17 / Python 2 |
|
| 0.17 / Python 3 | >=0.18 / any Python |
| 0.18 | >= 0.18 |
0.18.0 is backward-compatible for reading files packed by older versions, except for files packed with 0.17 in Python 2, in which case only they can only be unpacked in Python 2.
Signature change for .rank¶
Series.rank and DataFrame.rank now have the same signature (GH11759)
Previous signature
In [3]: pd.Series([0,1]).rank(method='average', na_option='keep',
ascending=True, pct=False)
Out[3]:
0 1
1 2
dtype: float64
In [4]: pd.DataFrame([0,1]).rank(axis=0, numeric_only=None,
method='average', na_option='keep',
ascending=True, pct=False)
Out[4]:
0
0 1
1 2
New signature
In [71]: pd.Series([0,1]).rank(axis=0, method='average', numeric_only=None,
....: na_option='keep', ascending=True, pct=False)
....:
Out[71]:
0 1.0
1 2.0
dtype: float64
In [72]: pd.DataFrame([0,1]).rank(axis=0, method='average', numeric_only=None,
....: na_option='keep', ascending=True, pct=False)
....:
�������������������������������������������Out[72]:
0
0 1.0
1 2.0
Bug in QuarterBegin with n=0¶
In previous versions, the behavior of the QuarterBegin offset was inconsistent
depending on the date when the n parameter was 0. (GH11406)
The general semantics of anchored offsets for n=0 is to not move the date
when it is an anchor point (e.g., a quarter start date), and otherwise roll
forward to the next anchor point.
In [73]: d = pd.Timestamp('2014-02-01')
In [74]: d
Out[74]: Timestamp('2014-02-01 00:00:00')
In [75]: d + pd.offsets.QuarterBegin(n=0, startingMonth=2)
������������������������������������������Out[75]: Timestamp('2014-02-01 00:00:00')
In [76]: d + pd.offsets.QuarterBegin(n=0, startingMonth=1)
������������������������������������������������������������������������������������Out[76]: Timestamp('2014-04-01 00:00:00')
For the QuarterBegin offset in previous versions, the date would be rolled
backwards if date was in the same month as the quarter start date.
In [3]: d = pd.Timestamp('2014-02-15')
In [4]: d + pd.offsets.QuarterBegin(n=0, startingMonth=2)
Out[4]: Timestamp('2014-02-01 00:00:00')
This behavior has been corrected in version 0.18.0, which is consistent with
other anchored offsets like MonthBegin and YearBegin.
In [77]: d = pd.Timestamp('2014-02-15')
In [78]: d + pd.offsets.QuarterBegin(n=0, startingMonth=2)
Out[78]: Timestamp('2014-05-01 00:00:00')
Resample API¶
Like the change in the window functions API above, .resample(...) is changing to have a more groupby-like API. (GH11732, GH12702, GH12202, GH12332, GH12334, GH12348, GH12448).
In [79]: np.random.seed(1234)
In [80]: df = pd.DataFrame(np.random.rand(10,4),
....: columns=list('ABCD'),
....: index=pd.date_range('2010-01-01 09:00:00', periods=10, freq='s'))
....:
In [81]: df
Out[81]:
A B C D
2010-01-01 09:00:00 0.191519 0.622109 0.437728 0.785359
2010-01-01 09:00:01 0.779976 0.272593 0.276464 0.801872
2010-01-01 09:00:02 0.958139 0.875933 0.357817 0.500995
2010-01-01 09:00:03 0.683463 0.712702 0.370251 0.561196
2010-01-01 09:00:04 0.503083 0.013768 0.772827 0.882641
2010-01-01 09:00:05 0.364886 0.615396 0.075381 0.368824
2010-01-01 09:00:06 0.933140 0.651378 0.397203 0.788730
2010-01-01 09:00:07 0.316836 0.568099 0.869127 0.436173
2010-01-01 09:00:08 0.802148 0.143767 0.704261 0.704581
2010-01-01 09:00:09 0.218792 0.924868 0.442141 0.909316
Previous API:
You would write a resampling operation that immediately evaluates. If a how parameter was not provided, it
would default to how='mean'.
In [6]: df.resample('2s')
Out[6]:
A B C D
2010-01-01 09:00:00 0.485748 0.447351 0.357096 0.793615
2010-01-01 09:00:02 0.820801 0.794317 0.364034 0.531096
2010-01-01 09:00:04 0.433985 0.314582 0.424104 0.625733
2010-01-01 09:00:06 0.624988 0.609738 0.633165 0.612452
2010-01-01 09:00:08 0.510470 0.534317 0.573201 0.806949
You could also specify a how directly
In [7]: df.resample('2s', how='sum')
Out[7]:
A B C D
2010-01-01 09:00:00 0.971495 0.894701 0.714192 1.587231
2010-01-01 09:00:02 1.641602 1.588635 0.728068 1.062191
2010-01-01 09:00:04 0.867969 0.629165 0.848208 1.251465
2010-01-01 09:00:06 1.249976 1.219477 1.266330 1.224904
2010-01-01 09:00:08 1.020940 1.068634 1.146402 1.613897
New API:
Now, you can write .resample(..) as a 2-stage operation like .groupby(...), which
yields a Resampler.
In [82]: r = df.resample('2s')
In [83]: r
Out[83]: DatetimeIndexResampler [freq=<2 * Seconds>, axis=0, closed=left, label=left, convention=start, base=0]
Downsampling¶
You can then use this object to perform operations. These are downsampling operations (going from a higher frequency to a lower one).
In [84]: r.mean()
Out[84]:
A B C D
2010-01-01 09:00:00 0.485748 0.447351 0.357096 0.793615
2010-01-01 09:00:02 0.820801 0.794317 0.364034 0.531096
2010-01-01 09:00:04 0.433985 0.314582 0.424104 0.625733
2010-01-01 09:00:06 0.624988 0.609738 0.633165 0.612452
2010-01-01 09:00:08 0.510470 0.534317 0.573201 0.806949
In [85]: r.sum()
Out[85]:
A B C D
2010-01-01 09:00:00 0.971495 0.894701 0.714192 1.587231
2010-01-01 09:00:02 1.641602 1.588635 0.728068 1.062191
2010-01-01 09:00:04 0.867969 0.629165 0.848208 1.251465
2010-01-01 09:00:06 1.249976 1.219477 1.266330 1.224904
2010-01-01 09:00:08 1.020940 1.068634 1.146402 1.613897
Furthermore, resample now supports getitem operations to perform the resample on specific columns.
In [86]: r[['A','C']].mean()
Out[86]:
A C
2010-01-01 09:00:00 0.485748 0.357096
2010-01-01 09:00:02 0.820801 0.364034
2010-01-01 09:00:04 0.433985 0.424104
2010-01-01 09:00:06 0.624988 0.633165
2010-01-01 09:00:08 0.510470 0.573201
and .aggregate type operations.
In [87]: r.agg({'A' : 'mean', 'B' : 'sum'})
Out[87]:
A B
2010-01-01 09:00:00 0.485748 0.894701
2010-01-01 09:00:02 0.820801 1.588635
2010-01-01 09:00:04 0.433985 0.629165
2010-01-01 09:00:06 0.624988 1.219477
2010-01-01 09:00:08 0.510470 1.068634
These accessors can of course, be combined
In [88]: r[['A','B']].agg(['mean','sum'])
Out[88]:
A B
mean sum mean sum
2010-01-01 09:00:00 0.485748 0.971495 0.447351 0.894701
2010-01-01 09:00:02 0.820801 1.641602 0.794317 1.588635
2010-01-01 09:00:04 0.433985 0.867969 0.314582 0.629165
2010-01-01 09:00:06 0.624988 1.249976 0.609738 1.219477
2010-01-01 09:00:08 0.510470 1.020940 0.534317 1.068634
Upsampling¶
Upsampling operations take you from a lower frequency to a higher frequency. These are now
performed with the Resampler objects with backfill(),
ffill(), fillna() and asfreq() methods.
In [89]: s = pd.Series(np.arange(5,dtype='int64'),
....: index=date_range('2010-01-01', periods=5, freq='Q'))
....:
In [90]: s
Out[90]:
2010-03-31 0
2010-06-30 1
2010-09-30 2
2010-12-31 3
2011-03-31 4
Freq: Q-DEC, dtype: int64
Previously
In [6]: s.resample('M', fill_method='ffill')
Out[6]:
2010-03-31 0
2010-04-30 0
2010-05-31 0
2010-06-30 1
2010-07-31 1
2010-08-31 1
2010-09-30 2
2010-10-31 2
2010-11-30 2
2010-12-31 3
2011-01-31 3
2011-02-28 3
2011-03-31 4
Freq: M, dtype: int64
New API
In [91]: s.resample('M').ffill()
Out[91]:
2010-03-31 0
2010-04-30 0
2010-05-31 0
2010-06-30 1
2010-07-31 1
2010-08-31 1
2010-09-30 2
2010-10-31 2
2010-11-30 2
2010-12-31 3
2011-01-31 3
2011-02-28 3
2011-03-31 4
Freq: M, dtype: int64
Note
In the new API, you can either downsample OR upsample. The prior implementation would allow you to pass an aggregator function (like mean) even though you were upsampling, providing a bit of confusion.
Previous API will work but with deprecations¶
Warning
This new API for resample includes some internal changes for the prior-to-0.18.0 API, to work with a deprecation warning in most cases, as the resample operation returns a deferred object. We can intercept operations and just do what the (pre 0.18.0) API did (with a warning). Here is a typical use case:
In [4]: r = df.resample('2s')
In [6]: r*10
pandas/tseries/resample.py:80: FutureWarning: .resample() is now a deferred operation
use .resample(...).mean() instead of .resample(...)
Out[6]:
A B C D
2010-01-01 09:00:00 4.857476 4.473507 3.570960 7.936154
2010-01-01 09:00:02 8.208011 7.943173 3.640340 5.310957
2010-01-01 09:00:04 4.339846 3.145823 4.241039 6.257326
2010-01-01 09:00:06 6.249881 6.097384 6.331650 6.124518
2010-01-01 09:00:08 5.104699 5.343172 5.732009 8.069486
However, getting and assignment operations directly on a Resampler will raise a ValueError:
In [7]: r.iloc[0] = 5
ValueError: .resample() is now a deferred operation
use .resample(...).mean() instead of .resample(...)
There is a situation where the new API can not perform all the operations when using original code.
This code is intending to resample every 2s, take the mean AND then take the min of those results.
In [4]: df.resample('2s').min()
Out[4]:
A 0.433985
B 0.314582
C 0.357096
D 0.531096
dtype: float64
The new API will:
In [92]: df.resample('2s').min()
Out[92]:
A B C D
2010-01-01 09:00:00 0.191519 0.272593 0.276464 0.785359
2010-01-01 09:00:02 0.683463 0.712702 0.357817 0.500995
2010-01-01 09:00:04 0.364886 0.013768 0.075381 0.368824
2010-01-01 09:00:06 0.316836 0.568099 0.397203 0.436173
2010-01-01 09:00:08 0.218792 0.143767 0.442141 0.704581
The good news is the return dimensions will differ between the new API and the old API, so this should loudly raise an exception.
To replicate the original operation
In [93]: df.resample('2s').mean().min()
Out[93]:
A 0.433985
B 0.314582
C 0.357096
D 0.531096
dtype: float64
Changes to eval¶
In prior versions, new columns assignments in an eval expression resulted
in an inplace change to the DataFrame. (GH9297, GH8664, GH10486)
In [94]: df = pd.DataFrame({'a': np.linspace(0, 10, 5), 'b': range(5)})
In [95]: df
Out[95]:
a b
0 0.0 0
1 2.5 1
2 5.0 2
3 7.5 3
4 10.0 4
In [12]: df.eval('c = a + b')
FutureWarning: eval expressions containing an assignment currentlydefault to operating inplace.
This will change in a future version of pandas, use inplace=True to avoid this warning.
In [13]: df
Out[13]:
a b c
0 0.0 0 0.0
1 2.5 1 3.5
2 5.0 2 7.0
3 7.5 3 10.5
4 10.0 4 14.0
In version 0.18.0, a new inplace keyword was added to choose whether the
assignment should be done inplace or return a copy.
In [96]: df
Out[96]:
a b c
0 0.0 0 0.0
1 2.5 1 3.5
2 5.0 2 7.0
3 7.5 3 10.5
4 10.0 4 14.0
In [97]: df.eval('d = c - b', inplace=False)
����������������������������������������������������������������������������������������������������������������Out[97]:
a b c d
0 0.0 0 0.0 0.0
1 2.5 1 3.5 2.5
2 5.0 2 7.0 5.0
3 7.5 3 10.5 7.5
4 10.0 4 14.0 10.0
In [98]: df
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[98]:
a b c
0 0.0 0 0.0
1 2.5 1 3.5
2 5.0 2 7.0
3 7.5 3 10.5
4 10.0 4 14.0
In [99]: df.eval('d = c - b', inplace=True)
In [100]: df
Out[100]:
a b c d
0 0.0 0 0.0 0.0
1 2.5 1 3.5 2.5
2 5.0 2 7.0 5.0
3 7.5 3 10.5 7.5
4 10.0 4 14.0 10.0
Warning
For backwards compatability, inplace defaults to True if not specified.
This will change in a future version of pandas. If your code depends on an
inplace assignment you should update to explicitly set inplace=True
The inplace keyword parameter was also added the query method.
In [101]: df.query('a > 5')
Out[101]:
a b c d
3 7.5 3 10.5 7.5
4 10.0 4 14.0 10.0
In [102]: df.query('a > 5', inplace=True)
In [103]: df
Out[103]:
a b c d
3 7.5 3 10.5 7.5
4 10.0 4 14.0 10.0
Warning
Note that the default value for inplace in a query
is False, which is consistent with prior versions.
eval has also been updated to allow multi-line expressions for multiple
assignments. These expressions will be evaluated one at a time in order. Only
assignments are valid for multi-line expressions.
In [104]: df
Out[104]:
a b c d
3 7.5 3 10.5 7.5
4 10.0 4 14.0 10.0
In [105]: df.eval("""
.....: e = d + a
.....: f = e - 22
.....: g = f / 2.0""", inplace=True)
.....:
In [106]: df
Out[106]:
a b c d e f g
3 7.5 3 10.5 7.5 15.0 -7.0 -3.5
4 10.0 4 14.0 10.0 20.0 -2.0 -1.0
Other API Changes¶
DataFrame.between_timeandSeries.between_timenow only parse a fixed set of time strings. Parsing of date strings is no longer supported and raises aValueError. (GH11818)In [107]: s = pd.Series(range(10), pd.date_range('2015-01-01', freq='H', periods=10)) In [108]: s.between_time("7:00am", "9:00am") Out[108]: 2015-01-01 07:00:00 7 2015-01-01 08:00:00 8 2015-01-01 09:00:00 9 Freq: H, dtype: int64
This will now raise.
In [2]: s.between_time('20150101 07:00:00','20150101 09:00:00') ValueError: Cannot convert arg ['20150101 07:00:00'] to a time.
.memory_usage()now includes values in the index, as does memory_usage in.info()(GH11597)DataFrame.to_latex()now supports non-ascii encodings (egutf-8) in Python 2 with the parameterencoding(GH7061)pandas.merge()andDataFrame.merge()will show a specific error message when trying to merge with an object that is not of typeDataFrameor a subclass (GH12081)DataFrame.unstackandSeries.unstacknow takefill_valuekeyword to allow direct replacement of missing values when an unstack results in missing values in the resultingDataFrame. As an added benefit, specifyingfill_valuewill preserve the data type of the original stacked data. (GH9746)As part of the new API for window functions and resampling, aggregation functions have been clarified, raising more informative error messages on invalid aggregations. (GH9052). A full set of examples are presented in groupby.
Statistical functions for
NDFrameobjects (likesum(), mean(), min()) will now raise if non-numpy-compatible arguments are passed in for**kwargs(GH12301).to_latexand.to_htmlgain adecimalparameter like.to_csv; the default is'.'(GH12031)More helpful error message when constructing a
DataFramewith empty data but with indices (GH8020).describe()will now properly handle bool dtype as a categorical (GH6625)More helpful error message with an invalid
.transformwith user defined input (GH10165)Exponentially weighted functions now allow specifying alpha directly (GH10789) and raise
ValueErrorif parameters violate0 < alpha <= 1(GH12492)
Deprecations¶
The functions
pd.rolling_*,pd.expanding_*, andpd.ewm*are deprecated and replaced by the corresponding method call. Note that the new suggested syntax includes all of the arguments (even if default) (GH11603)In [1]: s = pd.Series(range(3)) In [2]: pd.rolling_mean(s,window=2,min_periods=1) FutureWarning: pd.rolling_mean is deprecated for Series and will be removed in a future version, replace with Series.rolling(min_periods=1,window=2,center=False).mean() Out[2]: 0 0.0 1 0.5 2 1.5 dtype: float64 In [3]: pd.rolling_cov(s, s, window=2) FutureWarning: pd.rolling_cov is deprecated for Series and will be removed in a future version, replace with Series.rolling(window=2).cov(other=<Series>) Out[3]: 0 NaN 1 0.5 2 0.5 dtype: float64
The the
freqandhowarguments to the.rolling,.expanding, and.ewm(new) functions are deprecated, and will be removed in a future version. You can simply resample the input prior to creating a window function. (GH11603).For example, instead of
s.rolling(window=5,freq='D').max()to get the max value on a rolling 5 Day window, one could uses.resample('D').mean().rolling(window=5).max(), which first resamples the data to daily data, then provides a rolling 5 day window.pd.tseries.frequencies.get_offset_namefunction is deprecated. Use offset’s.freqstrproperty as alternative (GH11192)pandas.stats.fama_macbethroutines are deprecated and will be removed in a future version (GH6077)pandas.stats.ols,pandas.stats.plmandpandas.stats.varroutines are deprecated and will be removed in a future version (GH6077)show a
FutureWarningrather than aDeprecationWarningon using long-time deprecated syntax inHDFStore.select, where thewhereclause is not a string-like (GH12027)The
pandas.options.display.mpl_styleconfiguration has been deprecated and will be removed in a future version of pandas. This functionality is better handled by matplotlib’s style sheets (GH11783).
Removal of deprecated float indexers¶
In GH4892 indexing with floating point numbers on a non-Float64Index was deprecated (in version 0.14.0).
In 0.18.0, this deprecation warning is removed and these will now raise a TypeError. (GH12165, GH12333)
In [109]: s = pd.Series([1, 2, 3], index=[4, 5, 6])
In [110]: s
Out[110]:
4 1
5 2
6 3
dtype: int64
In [111]: s2 = pd.Series([1, 2, 3], index=list('abc'))
In [112]: s2
Out[112]:
a 1
b 2
c 3
dtype: int64
Previous Behavior:
# this is label indexing
In [2]: s[5.0]
FutureWarning: scalar indexers for index type Int64Index should be integers and not floating point
Out[2]: 2
# this is positional indexing
In [3]: s.iloc[1.0]
FutureWarning: scalar indexers for index type Int64Index should be integers and not floating point
Out[3]: 2
# this is label indexing
In [4]: s.loc[5.0]
FutureWarning: scalar indexers for index type Int64Index should be integers and not floating point
Out[4]: 2
# .ix would coerce 1.0 to the positional 1, and index
In [5]: s2.ix[1.0] = 10
FutureWarning: scalar indexers for index type Index should be integers and not floating point
In [6]: s2
Out[6]:
a 1
b 10
c 3
dtype: int64
New Behavior:
For iloc, getting & setting via a float scalar will always raise.
In [3]: s.iloc[2.0]
TypeError: cannot do label indexing on <class 'pandas.indexes.numeric.Int64Index'> with these indexers [2.0] of <type 'float'>
Other indexers will coerce to a like integer for both getting and setting. The FutureWarning has been dropped for .loc, .ix and [].
In [113]: s[5.0]
Out[113]: 2
In [114]: s.loc[5.0]
������������Out[114]: 2
and setting
In [115]: s_copy = s.copy()
In [116]: s_copy[5.0] = 10
In [117]: s_copy
Out[117]:
4 1
5 10
6 3
dtype: int64
In [118]: s_copy = s.copy()
In [119]: s_copy.loc[5.0] = 10
In [120]: s_copy
Out[120]:
4 1
5 10
6 3
dtype: int64
Positional setting with .ix and a float indexer will ADD this value to the index, rather than previously setting the value by position.
In [3]: s2.ix[1.0] = 10
In [4]: s2
Out[4]:
a 1
b 2
c 3
1.0 10
dtype: int64
Slicing will also coerce integer-like floats to integers for a non-Float64Index.
In [121]: s.loc[5.0:6]
Out[121]:
5 2
6 3
dtype: int64
Note that for floats that are NOT coercible to ints, the label based bounds will be excluded
In [122]: s.loc[5.1:6]
Out[122]:
6 3
dtype: int64
Float indexing on a Float64Index is unchanged.
In [123]: s = pd.Series([1, 2, 3], index=np.arange(3.))
In [124]: s[1.0]
Out[124]: 2
In [125]: s[1.0:2.5]
������������Out[125]:
1.0 2
2.0 3
dtype: int64
Removal of prior version deprecations/changes¶
- Removal of
rolling_corr_pairwisein favor of.rolling().corr(pairwise=True)(GH4950) - Removal of
expanding_corr_pairwisein favor of.expanding().corr(pairwise=True)(GH4950) - Removal of
DataMatrixmodule. This was not imported into the pandas namespace in any event (GH12111) - Removal of
colskeyword in favor ofsubsetinDataFrame.duplicated()andDataFrame.drop_duplicates()(GH6680) - Removal of the
read_frameandframe_query(both aliases forpd.read_sql) andwrite_frame(alias ofto_sql) functions in thepd.io.sqlnamespace, deprecated since 0.14.0 (GH6292). - Removal of the
orderkeyword from.factorize()(GH6930)
Performance Improvements¶
- Improved performance of
andrews_curves(GH11534) - Improved huge
DatetimeIndex,PeriodIndexandTimedeltaIndex‘s ops performance includingNaT(GH10277) - Improved performance of
pandas.concat(GH11958) - Improved performance of
StataReader(GH11591) - Improved performance in construction of
CategoricalswithSeriesof datetimes containingNaT(GH12077) - Improved performance of ISO 8601 date parsing for dates without separators (GH11899), leading zeros (GH11871) and with whitespace preceding the time zone (GH9714)
Bug Fixes¶
- Bug in
GroupBy.sizewhen data-frame is empty. (GH11699) - Bug in
Period.end_timewhen a multiple of time period is requested (GH11738) - Regression in
.clipwith tz-aware datetimes (GH11838) - Bug in
date_rangewhen the boundaries fell on the frequency (GH11804, GH12409) - Bug in consistency of passing nested dicts to
.groupby(...).agg(...)(GH9052) - Accept unicode in
Timedeltaconstructor (GH11995) - Bug in value label reading for
StataReaderwhen reading incrementally (GH12014) - Bug in vectorized
DateOffsetwhennparameter is0(GH11370) - Compat for numpy 1.11 w.r.t.
NaTcomparison changes (GH12049) - Bug in
read_csvwhen reading from aStringIOin threads (GH11790) - Bug in not treating
NaTas a missing value in datetimelikes when factorizing & withCategoricals(GH12077) - Bug in getitem when the values of a
Serieswere tz-aware (GH12089) - Bug in
Series.str.get_dummieswhen one of the variables was ‘name’ (GH12180) - Bug in
pd.concatwhile concatenating tz-aware NaT series. (GH11693, GH11755, GH12217) - Bug in
pd.read_statawith version <= 108 files (GH12232) - Bug in
Series.resampleusing a frequency ofNanowhen the index is aDatetimeIndexand contains non-zero nanosecond parts (GH12037) - Bug in resampling with
.nuniqueand a sparse index (GH12352) - Removed some compiler warnings (GH12471)
- Work around compat issues with
botoin python 3.5 (GH11915) - Bug in
NaTsubtraction fromTimestamporDatetimeIndexwith timezones (GH11718) - Bug in subtraction of
Seriesof a single tz-awareTimestamp(GH12290) - Use compat iterators in PY2 to support
.next()(GH12299) - Bug in
Timedelta.roundwith negative values (GH11690) - Bug in
.locagainstCategoricalIndexmay result in normalIndex(GH11586) - Bug in
DataFrame.infowhen duplicated column names exist (GH11761) - Bug in
.copyof datetime tz-aware objects (GH11794) - Bug in
Series.applyandSeries.mapwheretimedelta64was not boxed (GH11349) - Bug in
DataFrame.set_index()with tz-awareSeries(GH12358) - Bug in subclasses of
DataFramewhereAttributeErrordid not propagate (GH11808) - Bug groupby on tz-aware data where selection not returning
Timestamp(GH11616) - Bug in
pd.read_clipboardandpd.to_clipboardfunctions not supporting Unicode; upgrade includedpyperclipto v1.5.15 (GH9263) - Bug in
DataFrame.querycontaining an assignment (GH8664) - Bug in
from_msgpackwhere__contains__()fails for columns of the unpackedDataFrame, if theDataFramehas object columns. (GH11880) - Bug in
.resampleon categorical data withTimedeltaIndex(GH12169) - Bug in timezone info lost when broadcasting scalar datetime to
DataFrame(GH11682) - Bug in
Indexcreation fromTimestampwith mixed tz coerces to UTC (GH11488) - Bug in
to_numericwhere it does not raise if input is more than one dimension (GH11776) - Bug in parsing timezone offset strings with non-zero minutes (GH11708)
- Bug in
df.plotusing incorrect colors for bar plots under matplotlib 1.5+ (GH11614) - Bug in the
groupbyplotmethod when using keyword arguments (GH11805). - Bug in
DataFrame.duplicatedanddrop_duplicatescausing spurious matches when settingkeep=False(GH11864) - Bug in
.locresult with duplicated key may haveIndexwith incorrect dtype (GH11497) - Bug in
pd.rolling_medianwhere memory allocation failed even with sufficient memory (GH11696) - Bug in
DataFrame.stylewith spurious zeros (GH12134) - Bug in
DataFrame.stylewith integer columns not starting at 0 (GH12125) - Bug in
.style.barmay not rendered properly using specific browser (GH11678) - Bug in rich comparison of
Timedeltawith anumpy.arrayofTimedeltathat caused an infinite recursion (GH11835) - Bug in
DataFrame.rounddropping column index name (GH11986) - Bug in
df.replacewhile replacing value in mixed dtypeDataframe(GH11698) - Bug in
Indexprevents copying name of passedIndex, when a new name is not provided (GH11193) - Bug in
read_excelfailing to read any non-empty sheets when empty sheets exist andsheetname=None(GH11711) - Bug in
read_excelfailing to raiseNotImplementederror when keywordsparse_datesanddate_parserare provided (GH11544) - Bug in
read_sqlwithpymysqlconnections failing to return chunked data (GH11522) - Bug in
.to_csvignoring formatting parametersdecimal,na_rep,float_formatfor float indexes (GH11553) - Bug in
Int64IndexandFloat64Indexpreventing the use of the modulo operator (GH9244) - Bug in
MultiIndex.dropfor not lexsorted multi-indexes (GH12078) - Bug in
DataFramewhen masking an emptyDataFrame(GH11859) - Bug in
.plotpotentially modifying thecolorsinput when the number of columns didn’t match the number of series provided (GH12039). - Bug in
Series.plotfailing when index has aCustomBusinessDayfrequency (GH7222). - Bug in
.to_sqlfordatetime.timevalues with sqlite fallback (GH8341) - Bug in
read_excelfailing to read data with one column whensqueeze=True(GH12157) - Bug in
read_excelfailing to read one empty column (GH12292, GH9002) - Bug in
.groupbywhere aKeyErrorwas not raised for a wrong column if there was only one row in the dataframe (GH11741) - Bug in
.read_csvwith dtype specified on empty data producing an error (GH12048) - Bug in
.read_csvwhere strings like'2E'are treated as valid floats (GH12237) - Bug in building pandas with debugging symbols (GH12123)
- Removed
millisecondproperty ofDatetimeIndex. This would always raise aValueError(GH12019). - Bug in
Seriesconstructor with read-only data (GH11502) - Removed
pandas.util.testing.choice(). Should usenp.random.choice(), instead. (GH12386) - Bug in
.locsetitem indexer preventing the use of a TZ-aware DatetimeIndex (GH12050) - Bug in
.styleindexes and multi-indexes not appearing (GH11655) - Bug in
to_msgpackandfrom_msgpackwhich did not correctly serialize or deserializeNaT(GH12307). - Bug in
.skewand.kurtdue to roundoff error for highly similar values (GH11974) - Bug in
Timestampconstructor where microsecond resolution was lost if HHMMSS were not separated with ‘:’ (GH10041) - Bug in
buffer_rd_bytessrc->buffer could be freed more than once if reading failed, causing a segfault (GH12098) - Bug in
crosstabwhere arguments with non-overlapping indexes would return aKeyError(GH10291) - Bug in
DataFrame.applyin which reduction was not being prevented for cases in whichdtypewas not a numpy dtype (GH12244) - Bug when initializing categorical series with a scalar value. (GH12336)
- Bug when specifying a UTC
DatetimeIndexby settingutc=Truein.to_datetime(GH11934) - Bug when increasing the buffer size of CSV reader in
read_csv(GH12494) - Bug when setting columns of a
DataFramewith duplicate column names (GH12344)
v0.17.1 (November 21, 2015)¶
Note
We are proud to announce that pandas has become a sponsored project of the (NUMFocus organization). This will help ensure the success of development of pandas as a world-class open-source project.
This is a minor bug-fix release from 0.17.0 and includes a large number of bug fixes along several new features, enhancements, and performance improvements. We recommend that all users upgrade to this version.
Highlights include:
- Support for Conditional HTML Formatting, see here
- Releasing the GIL on the csv reader & other ops, see here
- Fixed regression in
DataFrame.drop_duplicatesfrom 0.16.2, causing incorrect results on integer values (GH11376)
What’s new in v0.17.1
New features¶
Conditional HTML Formatting¶
Warning
This is a new feature and is under active development. We’ll be adding features an possibly making breaking changes in future releases. Feedback is welcome.
We’ve added experimental support for conditional HTML formatting:
the visual styling of a DataFrame based on the data.
The styling is accomplished with HTML and CSS.
Acesses the styler class with the pandas.DataFrame.style, attribute,
an instance of Styler with your data attached.
Here’s a quick example:
In [1]: np.random.seed(123) In [2]: df = DataFrame(np.random.randn(10, 5), columns=list('abcde')) In [3]: html = df.style.background_gradient(cmap='viridis', low=.5)
We can render the HTML to get the following table.
| a | b | c | d | e | |
|---|---|---|---|---|---|
| 0 | -1.085631 | 0.997345 | 0.282978 | -1.506295 | -0.5786 |
| 1 | 1.651437 | -2.426679 | -0.428913 | 1.265936 | -0.86674 |
| 2 | -0.678886 | -0.094709 | 1.49139 | -0.638902 | -0.443982 |
| 3 | -0.434351 | 2.20593 | 2.186786 | 1.004054 | 0.386186 |
| 4 | 0.737369 | 1.490732 | -0.935834 | 1.175829 | -1.253881 |
| 5 | -0.637752 | 0.907105 | -1.428681 | -0.140069 | -0.861755 |
| 6 | -0.255619 | -2.798589 | -1.771533 | -0.699877 | 0.927462 |
| 7 | -0.173636 | 0.002846 | 0.688223 | -0.879536 | 0.283627 |
| 8 | -0.805367 | -1.727669 | -0.3909 | 0.573806 | 0.338589 |
| 9 | -0.01183 | 2.392365 | 0.412912 | 0.978736 | 2.238143 |
Styler interacts nicely with the Jupyter Notebook.
See the documentation for more.
Enhancements¶
DatetimeIndexnow supports conversion to strings withastype(str)(GH10442)Support for
compression(gzip/bz2) inpandas.DataFrame.to_csv()(GH7615)pd.read_*functions can now also acceptpathlib.Path, orpy._path.local.LocalPathobjects for thefilepath_or_bufferargument. (GH11033) - TheDataFrameandSeriesfunctions.to_csv(),.to_html()and.to_latex()can now handle paths beginning with tildes (e.g.~/Documents/) (GH11438)DataFramenow uses the fields of anamedtupleas columns, if columns are not supplied (GH11181)DataFrame.itertuples()now returnsnamedtupleobjects, when possible. (GH11269, GH11625)Added
axvlines_kwdsto parallel coordinates plot (GH10709)Option to
.info()and.memory_usage()to provide for deep introspection of memory consumption. Note that this can be expensive to compute and therefore is an optional parameter. (GH11595)In [4]: df = DataFrame({'A' : ['foo']*1000}) In [5]: df['B'] = df['A'].astype('category') # shows the '+' as we have object dtypes In [6]: df.info() <class 'pandas.core.frame.DataFrame'> RangeIndex: 1000 entries, 0 to 999 Data columns (total 2 columns): A 1000 non-null object B 1000 non-null category dtypes: category(1), object(1) memory usage: 9.0+ KB # we have an accurate memory assessment (but can be expensive to compute this) In [7]: df.info(memory_usage='deep') ��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������<class 'pandas.core.frame.DataFrame'> RangeIndex: 1000 entries, 0 to 999 Data columns (total 2 columns): A 1000 non-null object B 1000 non-null category dtypes: category(1), object(1) memory usage: 75.4 KB
Indexnow has afillnamethod (GH10089)In [8]: pd.Index([1, np.nan, 3]).fillna(2) Out[8]: Float64Index([1.0, 2.0, 3.0], dtype='float64')
Series of type
categorynow make.str.<...>and.dt.<...>accessor methods / properties available, if the categories are of that type. (GH10661)In [9]: s = pd.Series(list('aabb')).astype('category') In [10]: s Out[10]: 0 a 1 a 2 b 3 b dtype: category Categories (2, object): [a, b] In [11]: s.str.contains("a") �������������������������������������������������������������������������������������Out[11]: 0 True 1 True 2 False 3 False dtype: bool In [12]: date = pd.Series(pd.date_range('1/1/2015', periods=5)).astype('category') In [13]: date Out[13]: 0 2015-01-01 1 2015-01-02 2 2015-01-03 3 2015-01-04 4 2015-01-05 dtype: category Categories (5, datetime64[ns]): [2015-01-01, 2015-01-02, 2015-01-03, 2015-01-04, 2015-01-05] In [14]: date.dt.day ��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[14]: 0 1 1 2 2 3 3 4 4 5 dtype: int64
pivot_tablenow has amargins_nameargument so you can use something other than the default of ‘All’ (GH3335)Implement export of
datetime64[ns, tz]dtypes with a fixed HDF5 store (GH11411)Pretty printing sets (e.g. in DataFrame cells) now uses set literal syntax (
{x, y}) instead of Legacy Python syntax (set([x, y])) (GH11215)Improve the error message in
pandas.io.gbq.to_gbq()when a streaming insert fails (GH11285) and when the DataFrame does not match the schema of the destination table (GH11359)
API changes¶
- raise
NotImplementedErrorinIndex.shiftfor non-supported index types (GH8038) minandmaxreductions ondatetime64andtimedelta64dtyped series now result inNaTand notnan(GH11245).- Indexing with a null key will raise a
TypeError, instead of aValueError(GH11356) Series.ptpwill now ignore missing values by default (GH11163)
Performance Improvements¶
- Checking monotonic-ness before sorting on an index (GH11080)
Series.dropnaperformance improvement when its dtype can’t containNaN(GH11159)- Release the GIL on most datetime field operations (e.g.
DatetimeIndex.year,Series.dt.year), normalization, and conversion to and fromPeriod,DatetimeIndex.to_periodandPeriodIndex.to_timestamp(GH11263) - Release the GIL on some rolling algos:
rolling_median,rolling_mean,rolling_max,rolling_min,rolling_var,rolling_kurt,rolling_skew(GH11450) - Release the GIL when reading and parsing text files in
read_csv,read_table(GH11272) - Improved performance of
rolling_median(GH11450) - Improved performance of
to_excel(GH11352) - Performance bug in repr of
Categoricalcategories, which was rendering the strings before chopping them for display (GH11305) - Performance improvement in
Categorical.remove_unused_categories, (GH11643). - Improved performance of
Seriesconstructor with no data andDatetimeIndex(GH11433) - Improved performance of
shift,cumprod, andcumsumwith groupby (GH4095)
Bug Fixes¶
SparseArray.__iter__()now does not causePendingDeprecationWarningin Python 3.5 (GH11622)- Regression from 0.16.2 for output formatting of long floats/nan, restored in (GH11302)
Series.sort_index()now correctly handles theinplaceoption (GH11402)- Incorrectly distributed .c file in the build on
PyPiwhen reading a csv of floats and passingna_values=<a scalar>would show an exception (GH11374) - Bug in
.to_latex()output broken when the index has a name (GH10660) - Bug in
HDFStore.appendwith strings whose encoded length exceded the max unencoded length (GH11234) - Bug in merging
datetime64[ns, tz]dtypes (GH11405) - Bug in
HDFStore.selectwhen comparing with a numpy scalar in a where clause (GH11283) - Bug in using
DataFrame.ixwith a multi-index indexer (GH11372) - Bug in
date_rangewith ambigous endpoints (GH11626) - Prevent adding new attributes to the accessors
.str,.dtand.cat. Retrieving such a value was not possible, so error out on setting it. (GH10673) - Bug in tz-conversions with an ambiguous time and
.dtaccessors (GH11295) - Bug in output formatting when using an index of ambiguous times (GH11619)
- Bug in comparisons of Series vs list-likes (GH11339)
- Bug in
DataFrame.replacewith adatetime64[ns, tz]and a non-compat to_replace (GH11326, GH11153) - Bug in
isnullwherenumpy.datetime64('NaT')in anumpy.arraywas not determined to be null(GH11206) - Bug in list-like indexing with a mixed-integer Index (GH11320)
- Bug in
pivot_tablewithmargins=Truewhen indexes are ofCategoricaldtype (GH10993) - Bug in
DataFrame.plotcannot use hex strings colors (GH10299) - Regression in
DataFrame.drop_duplicatesfrom 0.16.2, causing incorrect results on integer values (GH11376) - Bug in
pd.evalwhere unary ops in a list error (GH11235) - Bug in
squeeze()with zero length arrays (GH11230, GH8999) - Bug in
describe()dropping column names for hierarchical indexes (GH11517) - Bug in
DataFrame.pct_change()not propagatingaxiskeyword on.fillnamethod (GH11150) - Bug in
.to_csv()when a mix of integer and string column names are passed as thecolumnsparameter (GH11637) - Bug in indexing with a
range, (GH11652) - Bug in inference of numpy scalars and preserving dtype when setting columns (GH11638)
- Bug in
to_sqlusing unicode column names giving UnicodeEncodeError with (GH11431). - Fix regression in setting of
xticksinplot(GH11529). - Bug in
holiday.dateswhere observance rules could not be applied to holiday and doc enhancement (GH11477, GH11533) - Fix plotting issues when having plain
Axesinstances instead ofSubplotAxes(GH11520, GH11556). - Bug in
DataFrame.to_latex()produces an extra rule whenheader=False(GH7124) - Bug in
df.groupby(...).apply(func)when a func returns aSeriescontaining a new datetimelike column (GH11324) - Bug in
pandas.jsonwhen file to load is big (GH11344) - Bugs in
to_excelwith duplicate columns (GH11007, GH10982, GH10970) - Fixed a bug that prevented the construction of an empty series of dtype
datetime64[ns, tz](GH11245). - Bug in
read_excelwith multi-index containing integers (GH11317) - Bug in
to_excelwith openpyxl 2.2+ and merging (GH11408) - Bug in
DataFrame.to_dict()produces anp.datetime64object instead ofTimestampwhen only datetime is present in data (GH11327) - Bug in
DataFrame.corr()raises exception when computes Kendall correlation for DataFrames with boolean and not boolean columns (GH11560) - Bug in the link-time error caused by C
inlinefunctions on FreeBSD 10+ (withclang) (GH10510) - Bug in
DataFrame.to_csvin passing through arguments for formattingMultiIndexes, includingdate_format(GH7791) - Bug in
DataFrame.join()withhow='right'producing aTypeError(GH11519) - Bug in
Series.quantilewith empty list results hasIndexwithobjectdtype (GH11588) - Bug in
pd.mergeresults in emptyInt64Indexrather thanIndex(dtype=object)when the merge result is empty (GH11588) - Bug in
Categorical.remove_unused_categorieswhen havingNaNvalues (GH11599) - Bug in
DataFrame.to_sparse()loses column names for MultiIndexes (GH11600) - Bug in
DataFrame.round()with non-unique column index producing a Fatal Python error (GH11611) - Bug in
DataFrame.round()withdecimalsbeing a non-unique indexed Series producing extra columns (GH11618)
v0.17.0 (October 9, 2015)¶
This is a major release from 0.16.2 and includes a small number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
Warning
pandas >= 0.17.0 will no longer support compatibility with Python version 3.2 (GH9118)
Warning
The pandas.io.data package is deprecated and will be replaced by the
pandas-datareader package.
This will allow the data modules to be independently updated to your pandas
installation. The API for pandas-datareader v0.1.1 is exactly the same
as in pandas v0.17.0 (GH8961, GH10861).
After installing pandas-datareader, you can easily change your imports:
from pandas.io import data, wb
becomes
from pandas_datareader import data, wb
Highlights include:
- Release the Global Interpreter Lock (GIL) on some cython operations, see here
- Plotting methods are now available as attributes of the
.plotaccessor, see here - The sorting API has been revamped to remove some long-time inconsistencies, see here
- Support for a
datetime64[ns]with timezones as a first-class dtype, see here - The default for
to_datetimewill now be toraisewhen presented with unparseable formats, previously this would return the original input. Also, date parse functions now return consistent results. See here - The default for
dropnainHDFStorehas changed toFalse, to store by default all rows even if they are allNaN, see here - Datetime accessor (
dt) now supportsSeries.dt.strftimeto generate formatted strings for datetime-likes, andSeries.dt.total_secondsto generate each duration of the timedelta in seconds. See here PeriodandPeriodIndexcan handle multiplied freq like3D, which corresponding to 3 days span. See here- Development installed versions of pandas will now have
PEP440compliant version strings (GH9518) - Development support for benchmarking with the Air Speed Velocity library (GH8361)
- Support for reading SAS xport files, see here
- Documentation comparing SAS to pandas, see here
- Removal of the automatic TimeSeries broadcasting, deprecated since 0.8.0, see here
- Display format with plain text can optionally align with Unicode East Asian Width, see here
- Compatibility with Python 3.5 (GH11097)
- Compatibility with matplotlib 1.5.0 (GH11111)
Check the API Changes and deprecations before updating.
What’s new in v0.17.0
- New features
- Datetime with TZ
- Releasing the GIL
- Plot submethods
- Additional methods for
dtaccessor - Period Frequency Enhancement
- Support for SAS XPORT files
- Support for Math Functions in .eval()
- Changes to Excel with
MultiIndex - Google BigQuery Enhancements
- Display Alignment with Unicode East Asian Width
- Other enhancements
- Backwards incompatible API changes
- Changes to sorting API
- Changes to to_datetime and to_timedelta
- Changes to Index Comparisons
- Changes to Boolean Comparisons vs. None
- HDFStore dropna behavior
- Changes to
display.precisionoption - Changes to
Categorical.unique - Changes to
boolpassed asheaderin Parsers - Other API Changes
- Deprecations
- Removal of prior version deprecations/changes
- Performance Improvements
- Bug Fixes
New features¶
Datetime with TZ¶
We are adding an implementation that natively supports datetime with timezones. A Series or a DataFrame column previously
could be assigned a datetime with timezones, and would work as an object dtype. This had performance issues with a large
number rows. See the docs for more details. (GH8260, GH10763, GH11034).
The new implementation allows for having a single-timezone across all rows, with operations in a performant manner.
In [1]: df = DataFrame({'A' : date_range('20130101',periods=3),
...: 'B' : date_range('20130101',periods=3,tz='US/Eastern'),
...: 'C' : date_range('20130101',periods=3,tz='CET')})
...:
In [2]: df
Out[2]:
A B C
0 2013-01-01 2013-01-01 00:00:00-05:00 2013-01-01 00:00:00+01:00
1 2013-01-02 2013-01-02 00:00:00-05:00 2013-01-02 00:00:00+01:00
2 2013-01-03 2013-01-03 00:00:00-05:00 2013-01-03 00:00:00+01:00
In [3]: df.dtypes
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[3]:
A datetime64[ns]
B datetime64[ns, US/Eastern]
C datetime64[ns, CET]
dtype: object
In [4]: df.B
Out[4]:
0 2013-01-01 00:00:00-05:00
1 2013-01-02 00:00:00-05:00
2 2013-01-03 00:00:00-05:00
Name: B, dtype: datetime64[ns, US/Eastern]
In [5]: df.B.dt.tz_localize(None)
����������������������������������������������������������������������������������������������������������������������������������������������Out[5]:
0 2013-01-01
1 2013-01-02
2 2013-01-03
Name: B, dtype: datetime64[ns]
This uses a new-dtype representation as well, that is very similar in look-and-feel to its numpy cousin datetime64[ns]
In [6]: df['B'].dtype
Out[6]: datetime64[ns, US/Eastern]
In [7]: type(df['B'].dtype)
�����������������������������������Out[7]: pandas.core.dtypes.dtypes.DatetimeTZDtype
Note
There is a slightly different string repr for the underlying DatetimeIndex as a result of the dtype changes, but
functionally these are the same.
Previous Behavior:
In [1]: pd.date_range('20130101',periods=3,tz='US/Eastern')
Out[1]: DatetimeIndex(['2013-01-01 00:00:00-05:00', '2013-01-02 00:00:00-05:00',
'2013-01-03 00:00:00-05:00'],
dtype='datetime64[ns]', freq='D', tz='US/Eastern')
In [2]: pd.date_range('20130101',periods=3,tz='US/Eastern').dtype
Out[2]: dtype('<M8[ns]')
New Behavior:
In [8]: pd.date_range('20130101',periods=3,tz='US/Eastern')
Out[8]:
DatetimeIndex(['2013-01-01 00:00:00-05:00', '2013-01-02 00:00:00-05:00',
'2013-01-03 00:00:00-05:00'],
dtype='datetime64[ns, US/Eastern]', freq='D')
In [9]: pd.date_range('20130101',periods=3,tz='US/Eastern').dtype
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[9]: datetime64[ns, US/Eastern]
Releasing the GIL¶
We are releasing the global-interpreter-lock (GIL) on some cython operations.
This will allow other threads to run simultaneously during computation, potentially allowing performance improvements
from multi-threading. Notably groupby, nsmallest, value_counts and some indexing operations benefit from this. (GH8882)
For example the groupby expression in the following code will have the GIL released during the factorization step, e.g. df.groupby('key')
as well as the .sum() operation.
N = 1000000
ngroups = 10
df = DataFrame({'key' : np.random.randint(0,ngroups,size=N),
'data' : np.random.randn(N) })
df.groupby('key')['data'].sum()
Releasing of the GIL could benefit an application that uses threads for user interactions (e.g. QT), or performing multi-threaded computations. A nice example of a library that can handle these types of computation-in-parallel is the dask library.
Plot submethods¶
The Series and DataFrame .plot() method allows for customizing plot types by supplying the kind keyword arguments. Unfortunately, many of these kinds of plots use different required and optional keyword arguments, which makes it difficult to discover what any given plot kind uses out of the dozens of possible arguments.
To alleviate this issue, we have added a new, optional plotting interface, which exposes each kind of plot as a method of the .plot attribute. Instead of writing series.plot(kind=<kind>, ...), you can now also use series.plot.<kind>(...):
In [10]: df = pd.DataFrame(np.random.rand(10, 2), columns=['a', 'b'])
In [11]: df.plot.bar()
As a result of this change, these methods are now all discoverable via tab-completion:
In [12]: df.plot.<TAB>
df.plot.area df.plot.barh df.plot.density df.plot.hist df.plot.line df.plot.scatter
df.plot.bar df.plot.box df.plot.hexbin df.plot.kde df.plot.pie
Each method signature only includes relevant arguments. Currently, these are limited to required arguments, but in the future these will include optional arguments, as well. For an overview, see the new Plotting API documentation.
Additional methods for dt accessor¶
strftime¶
We are now supporting a Series.dt.strftime method for datetime-likes to generate a formatted string (GH10110). Examples:
# DatetimeIndex
In [13]: s = pd.Series(pd.date_range('20130101', periods=4))
In [14]: s
Out[14]:
0 2013-01-01
1 2013-01-02
2 2013-01-03
3 2013-01-04
dtype: datetime64[ns]
In [15]: s.dt.strftime('%Y/%m/%d')
��������������������������������������������������������������������������������������������Out[15]:
0 2013/01/01
1 2013/01/02
2 2013/01/03
3 2013/01/04
dtype: object
# PeriodIndex
In [16]: s = pd.Series(pd.period_range('20130101', periods=4))
In [17]: s
Out[17]:
0 2013-01-01
1 2013-01-02
2 2013-01-03
3 2013-01-04
dtype: object
In [18]: s.dt.strftime('%Y/%m/%d')
������������������������������������������������������������������������������������Out[18]:
0 2013/01/01
1 2013/01/02
2 2013/01/03
3 2013/01/04
dtype: object
The string format is as the python standard library and details can be found here
total_seconds¶
pd.Series of type timedelta64 has new method .dt.total_seconds() returning the duration of the timedelta in seconds (GH10817)
# TimedeltaIndex
In [19]: s = pd.Series(pd.timedelta_range('1 minutes', periods=4))
In [20]: s
Out[20]:
0 0 days 00:01:00
1 1 days 00:01:00
2 2 days 00:01:00
3 3 days 00:01:00
dtype: timedelta64[ns]
In [21]: s.dt.total_seconds()
�����������������������������������������������������������������������������������������������������������������Out[21]:
0 60.0
1 86460.0
2 172860.0
3 259260.0
dtype: float64
Period Frequency Enhancement¶
Period, PeriodIndex and period_range can now accept multiplied freq. Also, Period.freq and PeriodIndex.freq are now stored as a DateOffset instance like DatetimeIndex, and not as str (GH7811)
A multiplied freq represents a span of corresponding length. The example below creates a period of 3 days. Addition and subtraction will shift the period by its span.
In [22]: p = pd.Period('2015-08-01', freq='3D')
In [23]: p
Out[23]: Period('2015-08-01', '3D')
In [24]: p + 1
������������������������������������Out[24]: Period('2015-08-04', '3D')
In [25]: p - 2
������������������������������������������������������������������������Out[25]: Period('2015-07-26', '3D')
In [26]: p.to_timestamp()
������������������������������������������������������������������������������������������������������������Out[26]: Timestamp('2015-08-01 00:00:00')
In [27]: p.to_timestamp(how='E')
������������������������������������������������������������������������������������������������������������������������������������������������������Out[27]: Timestamp('2015-08-03 00:00:00')
You can use the multiplied freq in PeriodIndex and period_range.
In [28]: idx = pd.period_range('2015-08-01', periods=4, freq='2D')
In [29]: idx
Out[29]: PeriodIndex(['2015-08-01', '2015-08-03', '2015-08-05', '2015-08-07'], dtype='period[2D]', freq='2D')
In [30]: idx + 1
��������������������������������������������������������������������������������������������������������������Out[30]: PeriodIndex(['2015-08-03', '2015-08-05', '2015-08-07', '2015-08-09'], dtype='period[2D]', freq='2D')
Support for SAS XPORT files¶
read_sas() provides support for reading SAS XPORT format files. (GH4052).
df = pd.read_sas('sas_xport.xpt')
It is also possible to obtain an iterator and read an XPORT file incrementally.
for df in pd.read_sas('sas_xport.xpt', chunksize=10000)
do_something(df)
See the docs for more details.
Support for Math Functions in .eval()¶
eval() now supports calling math functions (GH4893)
df = pd.DataFrame({'a': np.random.randn(10)})
df.eval("b = sin(a)")
The support math functions are sin, cos, exp, log, expm1, log1p, sqrt, sinh, cosh, tanh, arcsin, arccos, arctan, arccosh, arcsinh, arctanh, abs and arctan2.
These functions map to the intrinsics for the NumExpr engine. For the Python
engine, they are mapped to NumPy calls.
Changes to Excel with MultiIndex¶
In version 0.16.2 a DataFrame with MultiIndex columns could not be written to Excel via to_excel.
That functionality has been added (GH10564), along with updating read_excel so that the data can
be read back with, no loss of information, by specifying which columns/rows make up the MultiIndex
in the header and index_col parameters (GH4679)
See the documentation for more details.
In [31]: df = pd.DataFrame([[1,2,3,4], [5,6,7,8]],
....: columns = pd.MultiIndex.from_product([['foo','bar'],['a','b']],
....: names = ['col1', 'col2']),
....: index = pd.MultiIndex.from_product([['j'], ['l', 'k']],
....: names = ['i1', 'i2']))
....:
In [32]: df
Out[32]:
col1 foo bar
col2 a b a b
i1 i2
j l 1 2 3 4
k 5 6 7 8
In [33]: df.to_excel('test.xlsx')
In [34]: df = pd.read_excel('test.xlsx', header=[0,1], index_col=[0,1])
In [35]: df
Out[35]:
col1 foo bar
col2 a b a b
i1 i2
j l 1 2 3 4
k 5 6 7 8
Previously, it was necessary to specify the has_index_names argument in read_excel,
if the serialized data had index names. For version 0.17.0 the ouptput format of to_excel
has been changed to make this keyword unnecessary - the change is shown below.
Old
New
Warning
Excel files saved in version 0.16.2 or prior that had index names will still able to be read in,
but the has_index_names argument must specified to True.
Google BigQuery Enhancements¶
- Added ability to automatically create a table/dataset using the
pandas.io.gbq.to_gbq()function if the destination table/dataset does not exist. (GH8325, GH11121). - Added ability to replace an existing table and schema when calling the
pandas.io.gbq.to_gbq()function via theif_existsargument. See the docs for more details (GH8325). InvalidColumnOrderandInvalidPageTokenin the gbq module will raiseValueErrorinstead ofIOError.- The
generate_bq_schema()function is now deprecated and will be removed in a future version (GH11121) - The gbq module will now support Python 3 (GH11094).
Display Alignment with Unicode East Asian Width¶
Warning
Enabling this option will affect the performance for printing of DataFrame and Series (about 2 times slower).
Use only when it is actually required.
Some East Asian countries use Unicode characters its width is corresponding to 2 alphabets. If a DataFrame or Series contains these characters, the default output cannot be aligned properly. The following options are added to enable precise handling for these characters.
display.unicode.east_asian_width: Whether to use the Unicode East Asian Width to calculate the display text width. (GH2612)display.unicode.ambiguous_as_wide: Whether to handle Unicode characters belong to Ambiguous as Wide. (GH11102)
In [36]: df = pd.DataFrame({u'国籍': ['UK', u'日本'], u'名前': ['Alice', u'しのぶ']})
In [37]: df;

In [38]: pd.set_option('display.unicode.east_asian_width', True)
In [39]: df;
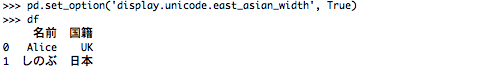
For further details, see here
Other enhancements¶
Support for
openpyxl>= 2.2. The API for style support is now stable (GH10125)mergenow accepts the argumentindicatorwhich adds a Categorical-type column (by default called_merge) to the output object that takes on the values (GH8790)Observation Origin _mergevalueMerge key only in 'left'frameleft_onlyMerge key only in 'right'frameright_onlyMerge key in both frames bothIn [40]: df1 = pd.DataFrame({'col1':[0,1], 'col_left':['a','b']}) In [41]: df2 = pd.DataFrame({'col1':[1,2,2],'col_right':[2,2,2]}) In [42]: pd.merge(df1, df2, on='col1', how='outer', indicator=True) Out[42]: col1 col_left col_right _merge 0 0 a NaN left_only 1 1 b 2.0 both 2 2 NaN 2.0 right_only 3 2 NaN 2.0 right_only
For more, see the updated docs
pd.to_numericis a new function to coerce strings to numbers (possibly with coercion) (GH11133)pd.mergewill now allow duplicate column names if they are not merged upon (GH10639).pd.pivotwill now allow passing index asNone(GH3962).pd.concatwill now use existing Series names if provided (GH10698).In [43]: foo = pd.Series([1,2], name='foo') In [44]: bar = pd.Series([1,2]) In [45]: baz = pd.Series([4,5])
Previous Behavior:
In [1] pd.concat([foo, bar, baz], 1) Out[1]: 0 1 2 0 1 1 4 1 2 2 5
New Behavior:
In [46]: pd.concat([foo, bar, baz], 1) Out[46]: foo 0 1 0 1 1 4 1 2 2 5
DataFramehas gained thenlargestandnsmallestmethods (GH10393)Add a
limit_directionkeyword argument that works withlimitto enableinterpolateto fillNaNvalues forward, backward, or both (GH9218, GH10420, GH11115)In [47]: ser = pd.Series([np.nan, np.nan, 5, np.nan, np.nan, np.nan, 13]) In [48]: ser.interpolate(limit=1, limit_direction='both') Out[48]: 0 NaN 1 5.0 2 5.0 3 7.0 4 NaN 5 11.0 6 13.0 dtype: float64
Added a
DataFrame.roundmethod to round the values to a variable number of decimal places (GH10568).In [49]: df = pd.DataFrame(np.random.random([3, 3]), columns=['A', 'B', 'C'], ....: index=['first', 'second', 'third']) ....: In [50]: df Out[50]: A B C first 0.342764 0.304121 0.417022 second 0.681301 0.875457 0.510422 third 0.669314 0.585937 0.624904 In [51]: df.round(2) ��������������������������������������������������������������������������������������������������������������������������������������������������������������Out[51]: A B C first 0.34 0.30 0.42 second 0.68 0.88 0.51 third 0.67 0.59 0.62 In [52]: df.round({'A': 0, 'C': 2}) ����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[52]: A B C first 0.0 0.304121 0.42 second 1.0 0.875457 0.51 third 1.0 0.585937 0.62
drop_duplicatesandduplicatednow accept akeepkeyword to target first, last, and all duplicates. Thetake_lastkeyword is deprecated, see here (GH6511, GH8505)In [53]: s = pd.Series(['A', 'B', 'C', 'A', 'B', 'D']) In [54]: s.drop_duplicates() Out[54]: 0 A 1 B 2 C 5 D dtype: object In [55]: s.drop_duplicates(keep='last') ����������������������������������������������������Out[55]: 2 C 3 A 4 B 5 D dtype: object In [56]: s.drop_duplicates(keep=False) ��������������������������������������������������������������������������������������������������������Out[56]: 2 C 5 D dtype: object
Reindex now has a
toleranceargument that allows for finer control of Limits on filling while reindexing (GH10411):In [57]: df = pd.DataFrame({'x': range(5), ....: 't': pd.date_range('2000-01-01', periods=5)}) ....: In [58]: df.reindex([0.1, 1.9, 3.5], ....: method='nearest', ....: tolerance=0.2) ....: Out[58]: t x 0.1 2000-01-01 0.0 1.9 2000-01-03 2.0 3.5 NaT NaN
When used on a
DatetimeIndex,TimedeltaIndexorPeriodIndex,tolerancewill coerced into aTimedeltaif possible. This allows you to specify tolerance with a string:In [59]: df = df.set_index('t') In [60]: df.reindex(pd.to_datetime(['1999-12-31']), ....: method='nearest', ....: tolerance='1 day') ....: Out[60]: x 1999-12-31 0
toleranceis also exposed by the lower levelIndex.get_indexerandIndex.get_locmethods.Added functionality to use the
baseargument when resampling aTimeDeltaIndex(GH10530)DatetimeIndexcan be instantiated using strings containsNaT(GH7599)to_datetimecan now accept theyearfirstkeyword (GH7599)pandas.tseries.offsetslarger than theDayoffset can now be used with aSeriesfor addition/subtraction (GH10699). See the docs for more details.pd.Timedelta.total_seconds()now returns Timedelta duration to ns precision (previously microsecond precision) (GH10939)PeriodIndexnow supports arithmetic withnp.ndarray(GH10638)Support pickling of
Periodobjects (GH10439).as_blockswill now take acopyoptional argument to return a copy of the data, default is to copy (no change in behavior from prior versions), (GH9607)regexargument toDataFrame.filternow handles numeric column names instead of raisingValueError(GH10384).Enable reading gzip compressed files via URL, either by explicitly setting the compression parameter or by inferring from the presence of the HTTP Content-Encoding header in the response (GH8685)
Enable writing Excel files in memory using StringIO/BytesIO (GH7074)
Enable serialization of lists and dicts to strings in
ExcelWriter(GH8188)SQL io functions now accept a SQLAlchemy connectable. (GH7877)
pd.read_sqlandto_sqlcan accept database URI asconparameter (GH10214)read_sql_tablewill now allow reading from views (GH10750).Enable writing complex values to
HDFStoreswhen using thetableformat (GH10447)Enable
pd.read_hdfto be used without specifying a key when the HDF file contains a single dataset (GH10443)pd.read_statawill now read Stata 118 type files. (GH9882)msgpacksubmodule has been updated to 0.4.6 with backward compatibility (GH10581)DataFrame.to_dictnow acceptsorient='index'keyword argument (GH10844).DataFrame.applywill return a Series of dicts if the passed function returns a dict andreduce=True(GH8735).Allow passing kwargs to the interpolation methods (GH10378).
Improved error message when concatenating an empty iterable of
Dataframeobjects (GH9157)pd.read_csvcan now read bz2-compressed files incrementally, and the C parser can read bz2-compressed files from AWS S3 (GH11070, GH11072).In
pd.read_csv, recognizes3n://ands3a://URLs as designating S3 file storage (GH11070, GH11071).Read CSV files from AWS S3 incrementally, instead of first downloading the entire file. (Full file download still required for compressed files in Python 2.) (GH11070, GH11073)
pd.read_csvis now able to infer compression type for files read from AWS S3 storage (GH11070, GH11074).
Backwards incompatible API changes¶
Changes to sorting API¶
The sorting API has had some longtime inconsistencies. (GH9816, GH8239).
Here is a summary of the API PRIOR to 0.17.0:
Series.sortis INPLACE whileDataFrame.sortreturns a new object.Series.orderreturns a new object- It was possible to use
Series/DataFrame.sort_indexto sort by values by passing thebykeyword. Series/DataFrame.sortlevelworked only on aMultiIndexfor sorting by index.
To address these issues, we have revamped the API:
- We have introduced a new method,
DataFrame.sort_values(), which is the merger ofDataFrame.sort(),Series.sort(), andSeries.order(), to handle sorting of values. - The existing methods
Series.sort(),Series.order(), andDataFrame.sort()have been deprecated and will be removed in a future version. - The
byargument ofDataFrame.sort_index()has been deprecated and will be removed in a future version. - The existing method
.sort_index()will gain thelevelkeyword to enable level sorting.
We now have two distinct and non-overlapping methods of sorting. A * marks items that
will show a FutureWarning.
To sort by the values:
| Previous | Replacement |
|---|---|
* Series.order() |
Series.sort_values() |
* Series.sort() |
Series.sort_values(inplace=True) |
* DataFrame.sort(columns=...) |
DataFrame.sort_values(by=...) |
To sort by the index:
| Previous | Replacement |
|---|---|
Series.sort_index() |
Series.sort_index() |
Series.sortlevel(level=...) |
Series.sort_index(level=...) |
DataFrame.sort_index() |
DataFrame.sort_index() |
DataFrame.sortlevel(level=...) |
DataFrame.sort_index(level=...) |
* DataFrame.sort() |
DataFrame.sort_index() |
We have also deprecated and changed similar methods in two Series-like classes, Index and Categorical.
| Previous | Replacement |
|---|---|
* Index.order() |
Index.sort_values() |
* Categorical.order() |
Categorical.sort_values() |
Changes to to_datetime and to_timedelta¶
Error handling¶
The default for pd.to_datetime error handling has changed to errors='raise'.
In prior versions it was errors='ignore'. Furthermore, the coerce argument
has been deprecated in favor of errors='coerce'. This means that invalid parsing
will raise rather that return the original input as in previous versions. (GH10636)
Previous Behavior:
In [2]: pd.to_datetime(['2009-07-31', 'asd'])
Out[2]: array(['2009-07-31', 'asd'], dtype=object)
New Behavior:
In [3]: pd.to_datetime(['2009-07-31', 'asd'])
ValueError: Unknown string format
Of course you can coerce this as well.
In [61]: to_datetime(['2009-07-31', 'asd'], errors='coerce')
Out[61]: DatetimeIndex(['2009-07-31', 'NaT'], dtype='datetime64[ns]', freq=None)
To keep the previous behavior, you can use errors='ignore':
In [62]: to_datetime(['2009-07-31', 'asd'], errors='ignore')
Out[62]: array(['2009-07-31', 'asd'], dtype=object)
Furthermore, pd.to_timedelta has gained a similar API, of errors='raise'|'ignore'|'coerce', and the coerce keyword
has been deprecated in favor of errors='coerce'.
Consistent Parsing¶
The string parsing of to_datetime, Timestamp and DatetimeIndex has
been made consistent. (GH7599)
Prior to v0.17.0, Timestamp and to_datetime may parse year-only datetime-string incorrectly using today’s date, otherwise DatetimeIndex
uses the beginning of the year. Timestamp and to_datetime may raise ValueError in some types of datetime-string which DatetimeIndex
can parse, such as a quarterly string.
Previous Behavior:
In [1]: Timestamp('2012Q2')
Traceback
...
ValueError: Unable to parse 2012Q2
# Results in today's date.
In [2]: Timestamp('2014')
Out [2]: 2014-08-12 00:00:00
v0.17.0 can parse them as below. It works on DatetimeIndex also.
New Behavior:
In [63]: Timestamp('2012Q2')
Out[63]: Timestamp('2012-04-01 00:00:00')
In [64]: Timestamp('2014')
������������������������������������������Out[64]: Timestamp('2014-01-01 00:00:00')
In [65]: DatetimeIndex(['2012Q2', '2014'])
������������������������������������������������������������������������������������Out[65]: DatetimeIndex(['2012-04-01', '2014-01-01'], dtype='datetime64[ns]', freq=None)
Note
If you want to perform calculations based on today’s date, use Timestamp.now() and pandas.tseries.offsets.
In [66]: import pandas.tseries.offsets as offsets
In [67]: Timestamp.now()
Out[67]: Timestamp('2017-07-07 12:29:28.795000')
In [68]: Timestamp.now() + offsets.DateOffset(years=1)
�������������������������������������������������Out[68]: Timestamp('2018-07-07 12:29:28.796446')
Changes to Index Comparisons¶
Operator equal on Index should behavior similarly to Series (GH9947, GH10637)
Starting in v0.17.0, comparing Index objects of different lengths will raise
a ValueError. This is to be consistent with the behavior of Series.
Previous Behavior:
In [2]: pd.Index([1, 2, 3]) == pd.Index([1, 4, 5])
Out[2]: array([ True, False, False], dtype=bool)
In [3]: pd.Index([1, 2, 3]) == pd.Index([2])
Out[3]: array([False, True, False], dtype=bool)
In [4]: pd.Index([1, 2, 3]) == pd.Index([1, 2])
Out[4]: False
New Behavior:
In [8]: pd.Index([1, 2, 3]) == pd.Index([1, 4, 5])
Out[8]: array([ True, False, False], dtype=bool)
In [9]: pd.Index([1, 2, 3]) == pd.Index([2])
ValueError: Lengths must match to compare
In [10]: pd.Index([1, 2, 3]) == pd.Index([1, 2])
ValueError: Lengths must match to compare
Note that this is different from the numpy behavior where a comparison can
be broadcast:
In [69]: np.array([1, 2, 3]) == np.array([1])
Out[69]: array([ True, False, False], dtype=bool)
or it can return False if broadcasting can not be done:
In [70]: np.array([1, 2, 3]) == np.array([1, 2])
Out[70]: False
Changes to Boolean Comparisons vs. None¶
Boolean comparisons of a Series vs None will now be equivalent to comparing with np.nan, rather than raise TypeError. (GH1079).
In [71]: s = Series(range(3))
In [72]: s.iloc[1] = None
In [73]: s
Out[73]:
0 0.0
1 NaN
2 2.0
dtype: float64
Previous Behavior:
In [5]: s==None
TypeError: Could not compare <type 'NoneType'> type with Series
New Behavior:
In [74]: s==None
Out[74]:
0 False
1 False
2 False
dtype: bool
Usually you simply want to know which values are null.
In [75]: s.isnull()
Out[75]:
0 False
1 True
2 False
dtype: bool
Warning
You generally will want to use isnull/notnull for these types of comparisons, as isnull/notnull tells you which elements are null. One has to be
mindful that nan's don’t compare equal, but None's do. Note that Pandas/numpy uses the fact that np.nan != np.nan, and treats None like np.nan.
In [76]: None == None
Out[76]: True
In [77]: np.nan == np.nan
��������������Out[77]: False
HDFStore dropna behavior¶
The default behavior for HDFStore write functions with format='table' is now to keep rows that are all missing. Previously, the behavior was to drop rows that were all missing save the index. The previous behavior can be replicated using the dropna=True option. (GH9382)
Previous Behavior:
In [78]: df_with_missing = pd.DataFrame({'col1':[0, np.nan, 2],
....: 'col2':[1, np.nan, np.nan]})
....:
In [79]: df_with_missing
Out[79]:
col1 col2
0 0.0 1.0
1 NaN NaN
2 2.0 NaN
In [27]:
df_with_missing.to_hdf('file.h5',
'df_with_missing',
format='table',
mode='w')
In [28]: pd.read_hdf('file.h5', 'df_with_missing')
Out [28]:
col1 col2
0 0 1
2 2 NaN
New Behavior:
In [80]: df_with_missing.to_hdf('file.h5',
....: 'df_with_missing',
....: format='table',
....: mode='w')
....:
In [81]: pd.read_hdf('file.h5', 'df_with_missing')
Out[81]:
col1 col2
0 0.0 1.0
1 NaN NaN
2 2.0 NaN
See the docs for more details.
Changes to display.precision option¶
The display.precision option has been clarified to refer to decimal places (GH10451).
Earlier versions of pandas would format floating point numbers to have one less decimal place than the value in
display.precision.
In [1]: pd.set_option('display.precision', 2)
In [2]: pd.DataFrame({'x': [123.456789]})
Out[2]:
x
0 123.5
If interpreting precision as “significant figures” this did work for scientific notation but that same interpretation did not work for values with standard formatting. It was also out of step with how numpy handles formatting.
Going forward the value of display.precision will directly control the number of places after the decimal, for
regular formatting as well as scientific notation, similar to how numpy’s precision print option works.
In [82]: pd.set_option('display.precision', 2)
In [83]: pd.DataFrame({'x': [123.456789]})
Out[83]:
x
0 123.46
To preserve output behavior with prior versions the default value of display.precision has been reduced to 6
from 7.
Changes to Categorical.unique¶
Categorical.unique now returns new Categoricals with categories and codes that are unique, rather than returning np.array (GH10508)
- unordered category: values and categories are sorted by appearance order.
- ordered category: values are sorted by appearance order, categories keep existing order.
In [84]: cat = pd.Categorical(['C', 'A', 'B', 'C'],
....: categories=['A', 'B', 'C'],
....: ordered=True)
....:
In [85]: cat
Out[85]:
[C, A, B, C]
Categories (3, object): [A < B < C]
In [86]: cat.unique()
�����������������������������������������������������������Out[86]:
[C, A, B]
Categories (3, object): [A < B < C]
In [87]: cat = pd.Categorical(['C', 'A', 'B', 'C'],
....: categories=['A', 'B', 'C'])
....:
In [88]: cat
Out[88]:
[C, A, B, C]
Categories (3, object): [A, B, C]
In [89]: cat.unique()
���������������������������������������������������������Out[89]:
[C, A, B]
Categories (3, object): [C, A, B]
Changes to bool passed as header in Parsers¶
In earlier versions of pandas, if a bool was passed the header argument of
read_csv, read_excel, or read_html it was implicitly converted to
an integer, resulting in header=0 for False and header=1 for True
(GH6113)
A bool input to header will now raise a TypeError
In [29]: df = pd.read_csv('data.csv', header=False)
TypeError: Passing a bool to header is invalid. Use header=None for no header or
header=int or list-like of ints to specify the row(s) making up the column names
Other API Changes¶
Line and kde plot with
subplots=Truenow uses default colors, not all black. Specifycolor='k'to draw all lines in black (GH9894)Calling the
.value_counts()method on a Series with acategoricaldtype now returns a Series with aCategoricalIndex(GH10704)The metadata properties of subclasses of pandas objects will now be serialized (GH10553).
groupbyusingCategoricalfollows the same rule asCategorical.uniquedescribed above (GH10508)When constructing
DataFramewith an array ofcomplex64dtype previously meant the corresponding column was automatically promoted to thecomplex128dtype. Pandas will now preserve the itemsize of the input for complex data (GH10952)some numeric reduction operators would return
ValueError, rather thanTypeErroron object types that includes strings and numbers (GH11131)Passing currently unsupported
chunksizeargument toread_excelorExcelFile.parsewill now raiseNotImplementedError(GH8011)Allow an
ExcelFileobject to be passed intoread_excel(GH11198)DatetimeIndex.uniondoes not inferfreqifselfand the input haveNoneasfreq(GH11086)NaT‘s methods now either raiseValueError, or returnnp.nanorNaT(GH9513)Behavior Methods return np.nanweekday,isoweekdayreturn NaTdate,now,replace,to_datetime,todayreturn np.datetime64('NaT')to_datetime64(unchanged)raise ValueErrorAll other public methods (names not beginning with underscores)
Deprecations¶
For
Seriesthe following indexing functions are deprecated (GH10177).Deprecated Function Replacement .irow(i).iloc[i]or.iat[i].iget(i).iloc[i]or.iat[i].iget_value(i).iloc[i]or.iat[i]For
DataFramethe following indexing functions are deprecated (GH10177).Deprecated Function Replacement .irow(i).iloc[i].iget_value(i, j).iloc[i, j]or.iat[i, j].icol(j).iloc[:, j]
Note
These indexing function have been deprecated in the documentation since 0.11.0.
Categorical.namewas deprecated to makeCategoricalmorenumpy.ndarraylike. UseSeries(cat, name="whatever")instead (GH10482).- Setting missing values (NaN) in a
Categorical‘scategorieswill issue a warning (GH10748). You can still have missing values in thevalues. drop_duplicatesandduplicated‘stake_lastkeyword was deprecated in favor ofkeep. (GH6511, GH8505)Series.nsmallestandnlargest‘stake_lastkeyword was deprecated in favor ofkeep. (GH10792)DataFrame.combineAddandDataFrame.combineMultare deprecated. They can easily be replaced by using theaddandmulmethods:DataFrame.add(other, fill_value=0)andDataFrame.mul(other, fill_value=1.)(GH10735).TimeSeriesdeprecated in favor ofSeries(note that this has been an alias since 0.13.0), (GH10890)SparsePaneldeprecated and will be removed in a future version (GH11157).Series.is_time_seriesdeprecated in favor ofSeries.index.is_all_dates(GH11135)- Legacy offsets (like
'A@JAN') are deprecated (note that this has been alias since 0.8.0) (GH10878) WidePaneldeprecated in favor ofPanel,LongPanelin favor ofDataFrame(note these have been aliases since < 0.11.0), (GH10892)DataFrame.convert_objectshas been deprecated in favor of type-specific functionspd.to_datetime,pd.to_timestampandpd.to_numeric(new in 0.17.0) (GH11133).
Removal of prior version deprecations/changes¶
Removal of
na_lastparameters fromSeries.order()andSeries.sort(), in favor ofna_position. (GH5231)Remove of
percentile_widthfrom.describe(), in favor ofpercentiles. (GH7088)Removal of
colSpaceparameter fromDataFrame.to_string(), in favor ofcol_space, circa 0.8.0 version.Removal of automatic time-series broadcasting (GH2304)
In [90]: np.random.seed(1234) In [91]: df = DataFrame(np.random.randn(5,2),columns=list('AB'),index=date_range('20130101',periods=5)) In [92]: df Out[92]: A B 2013-01-01 0.471435 -1.190976 2013-01-02 1.432707 -0.312652 2013-01-03 -0.720589 0.887163 2013-01-04 0.859588 -0.636524 2013-01-05 0.015696 -2.242685
Previously
In [3]: df + df.A FutureWarning: TimeSeries broadcasting along DataFrame index by default is deprecated. Please use DataFrame.<op> to explicitly broadcast arithmetic operations along the index Out[3]: A B 2013-01-01 0.942870 -0.719541 2013-01-02 2.865414 1.120055 2013-01-03 -1.441177 0.166574 2013-01-04 1.719177 0.223065 2013-01-05 0.031393 -2.226989
Current
In [93]: df.add(df.A,axis='index') Out[93]: A B 2013-01-01 0.942870 -0.719541 2013-01-02 2.865414 1.120055 2013-01-03 -1.441177 0.166574 2013-01-04 1.719177 0.223065 2013-01-05 0.031393 -2.226989
Remove
tablekeyword inHDFStore.put/append, in favor of usingformat=(GH4645)Remove
kindinread_excel/ExcelFileas its unused (GH4712)Remove
infer_typekeyword frompd.read_htmlas its unused (GH4770, GH7032)Remove
offsetandtimeRulekeywords fromSeries.tshift/shift, in favor offreq(GH4853, GH4864)Remove
pd.load/pd.savealiases in favor ofpd.to_pickle/pd.read_pickle(GH3787)
Performance Improvements¶
- Development support for benchmarking with the Air Speed Velocity library (GH8361)
- Added vbench benchmarks for alternative ExcelWriter engines and reading Excel files (GH7171)
- Performance improvements in
Categorical.value_counts(GH10804) - Performance improvements in
SeriesGroupBy.nuniqueandSeriesGroupBy.value_countsandSeriesGroupby.transform(GH10820, GH11077) - Performance improvements in
DataFrame.drop_duplicateswith integer dtypes (GH10917) - Performance improvements in
DataFrame.duplicatedwith wide frames. (GH10161, GH11180) - 4x improvement in
timedeltastring parsing (GH6755, GH10426) - 8x improvement in
timedelta64anddatetime64ops (GH6755) - Significantly improved performance of indexing
MultiIndexwith slicers (GH10287) - 8x improvement in
ilocusing list-like input (GH10791) - Improved performance of
Series.isinfor datetimelike/integer Series (GH10287) - 20x improvement in
concatof Categoricals when categories are identical (GH10587) - Improved performance of
to_datetimewhen specified format string is ISO8601 (GH10178) - 2x improvement of
Series.value_countsfor float dtype (GH10821) - Enable
infer_datetime_formatinto_datetimewhen date components do not have 0 padding (GH11142) - Regression from 0.16.1 in constructing
DataFramefrom nested dictionary (GH11084) - Performance improvements in addition/subtraction operations for
DateOffsetwithSeriesorDatetimeIndex(GH10744, GH11205)
Bug Fixes¶
- Bug in incorrection computation of
.mean()ontimedelta64[ns]because of overflow (GH9442) - Bug in
.isinon older numpies (:issue: 11232) - Bug in
DataFrame.to_html(index=False)renders unnecessarynamerow (GH10344) - Bug in
DataFrame.to_latex()thecolumn_formatargument could not be passed (GH9402) - Bug in
DatetimeIndexwhen localizing withNaT(GH10477) - Bug in
Series.dtops in preserving meta-data (GH10477) - Bug in preserving
NaTwhen passed in an otherwise invalidto_datetimeconstruction (GH10477) - Bug in
DataFrame.applywhen function returns categorical series. (GH9573) - Bug in
to_datetimewith invalid dates and formats supplied (GH10154) - Bug in
Index.drop_duplicatesdropping name(s) (GH10115) - Bug in
Series.quantiledropping name (GH10881) - Bug in
pd.Serieswhen setting a value on an emptySerieswhose index has a frequency. (GH10193) - Bug in
pd.Series.interpolatewith invalidorderkeyword values. (GH10633) - Bug in
DataFrame.plotraisesValueErrorwhen color name is specified by multiple characters (GH10387) - Bug in
Indexconstruction with a mixed list of tuples (GH10697) - Bug in
DataFrame.reset_indexwhen index containsNaT. (GH10388) - Bug in
ExcelReaderwhen worksheet is empty (GH6403) - Bug in
BinGrouper.group_infowhere returned values are not compatible with base class (GH10914) - Bug in clearing the cache on
DataFrame.popand a subsequent inplace op (GH10912) - Bug in indexing with a mixed-integer
Indexcausing anImportError(GH10610) - Bug in
Series.countwhen index has nulls (GH10946) - Bug in pickling of a non-regular freq
DatetimeIndex(GH11002) - Bug causing
DataFrame.whereto not respect theaxisparameter when the frame has a symmetric shape. (GH9736) - Bug in
Table.select_columnwhere name is not preserved (GH10392) - Bug in
offsets.generate_rangewherestartandendhave finer precision thanoffset(GH9907) - Bug in
pd.rolling_*whereSeries.namewould be lost in the output (GH10565) - Bug in
stackwhen index or columns are not unique. (GH10417) - Bug in setting a
Panelwhen an axis has a multi-index (GH10360) - Bug in
USFederalHolidayCalendarwhereUSMemorialDayandUSMartinLutherKingJrwere incorrect (GH10278 and GH9760 ) - Bug in
.sample()where returned object, if set, gives unnecessarySettingWithCopyWarning(GH10738) - Bug in
.sample()where weights passed asSerieswere not aligned along axis before being treated positionally, potentially causing problems if weight indices were not aligned with sampled object. (GH10738) - Regression fixed in (GH9311, GH6620, GH9345), where groupby with a datetime-like converting to float with certain aggregators (GH10979)
- Bug in
DataFrame.interpolatewithaxis=1andinplace=True(GH10395) - Bug in
io.sql.get_schemawhen specifying multiple columns as primary key (GH10385). - Bug in
groupby(sort=False)with datetime-likeCategoricalraisesValueError(GH10505) - Bug in
groupby(axis=1)withfilter()throwsIndexError(GH11041) - Bug in
test_categoricalon big-endian builds (GH10425) - Bug in
Series.shiftandDataFrame.shiftnot supporting categorical data (GH9416) - Bug in
Series.mapusing categoricalSeriesraisesAttributeError(GH10324) - Bug in
MultiIndex.get_level_valuesincludingCategoricalraisesAttributeError(GH10460) - Bug in
pd.get_dummieswithsparse=Truenot returningSparseDataFrame(GH10531) - Bug in
Indexsubtypes (such asPeriodIndex) not returning their own type for.dropand.insertmethods (GH10620) - Bug in
algos.outer_join_indexerwhenrightarray is empty (GH10618) - Bug in
filter(regression from 0.16.0) andtransformwhen grouping on multiple keys, one of which is datetime-like (GH10114) - Bug in
to_datetimeandto_timedeltacausingIndexname to be lost (GH10875) - Bug in
len(DataFrame.groupby)causingIndexErrorwhen there’s a column containing only NaNs (:issue: 11016) - Bug that caused segfault when resampling an empty Series (GH10228)
- Bug in
DatetimeIndexandPeriodIndex.value_countsresets name from its result, but retains in result’sIndex. (GH10150) - Bug in
pd.evalusingnumexprengine coerces 1 element numpy array to scalar (GH10546) - Bug in
pd.concatwithaxis=0when column is of dtypecategory(GH10177) - Bug in
read_msgpackwhere input type is not always checked (GH10369, GH10630) - Bug in
pd.read_csvwith kwargsindex_col=False,index_col=['a', 'b']ordtype(GH10413, GH10467, GH10577) - Bug in
Series.from_csvwithheaderkwarg not setting theSeries.nameor theSeries.index.name(GH10483) - Bug in
groupby.varwhich caused variance to be inaccurate for small float values (GH10448) - Bug in
Series.plot(kind='hist')Y Label not informative (GH10485) - Bug in
read_csvwhen using a converter which generates auint8type (GH9266) - Bug causes memory leak in time-series line and area plot (GH9003)
- Bug when setting a
Panelsliced along the major or minor axes when the right-hand side is aDataFrame(GH11014) - Bug that returns
Noneand does not raiseNotImplementedErrorwhen operator functions (e.g..add) ofPanelare not implemented (GH7692) - Bug in line and kde plot cannot accept multiple colors when
subplots=True(GH9894) - Bug in
DataFrame.plotraisesValueErrorwhen color name is specified by multiple characters (GH10387) - Bug in left and right
alignofSerieswithMultiIndexmay be inverted (GH10665) - Bug in left and right
joinof withMultiIndexmay be inverted (GH10741) - Bug in
read_statawhen reading a file with a different order set incolumns(GH10757) - Bug in
Categoricalmay not representing properly when category containstzorPeriod(GH10713) - Bug in
Categorical.__iter__may not returning correctdatetimeandPeriod(GH10713) - Bug in indexing with a
PeriodIndexon an object with aPeriodIndex(GH4125) - Bug in
read_csvwithengine='c': EOF preceded by a comment, blank line, etc. was not handled correctly (GH10728, GH10548) - Reading “famafrench” data via
DataReaderresults in HTTP 404 error because of the website url is changed (GH10591). - Bug in
read_msgpackwhere DataFrame to decode has duplicate column names (GH9618) - Bug in
io.common.get_filepath_or_bufferwhich caused reading of valid S3 files to fail if the bucket also contained keys for which the user does not have read permission (GH10604) - Bug in vectorised setting of timestamp columns with python
datetime.dateand numpydatetime64(GH10408, GH10412) - Bug in
Index.takemay add unnecessaryfreqattribute (GH10791) - Bug in
mergewith emptyDataFramemay raiseIndexError(GH10824) - Bug in
to_latexwhere unexpected keyword argument for some documented arguments (GH10888) - Bug in indexing of large
DataFramewhereIndexErroris uncaught (GH10645 and GH10692) - Bug in
read_csvwhen using thenrowsorchunksizeparameters if file contains only a header line (GH9535) - Bug in serialization of
categorytypes in HDF5 in presence of alternate encodings. (GH10366) - Bug in
pd.DataFramewhen constructing an empty DataFrame with a string dtype (GH9428) - Bug in
pd.DataFrame.diffwhen DataFrame is not consolidated (GH10907) - Bug in
pd.uniquefor arrays with thedatetime64ortimedelta64dtype that meant an array with object dtype was returned instead the original dtype (GH9431) - Bug in
Timedeltaraising error when slicing from 0s (GH10583) - Bug in
DatetimeIndex.takeandTimedeltaIndex.takemay not raiseIndexErroragainst invalid index (GH10295) - Bug in
Series([np.nan]).astype('M8[ms]'), which now returnsSeries([pd.NaT])(GH10747) - Bug in
PeriodIndex.orderreset freq (GH10295) - Bug in
date_rangewhenfreqdividesendas nanos (GH10885) - Bug in
ilocallowing memory outside bounds of a Series to be accessed with negative integers (GH10779) - Bug in
read_msgpackwhere encoding is not respected (GH10581) - Bug preventing access to the first index when using
ilocwith a list containing the appropriate negative integer (GH10547, GH10779) - Bug in
TimedeltaIndexformatter causing error while trying to saveDataFramewithTimedeltaIndexusingto_csv(GH10833) - Bug in
DataFrame.wherewhen handling Series slicing (GH10218, GH9558) - Bug where
pd.read_gbqthrowsValueErrorwhen Bigquery returns zero rows (GH10273) - Bug in
to_jsonwhich was causing segmentation fault when serializing 0-rank ndarray (GH9576) - Bug in plotting functions may raise
IndexErrorwhen plotted onGridSpec(GH10819) - Bug in plot result may show unnecessary minor ticklabels (GH10657)
- Bug in
groupbyincorrect computation for aggregation onDataFramewithNaT(E.gfirst,last,min). (GH10590, GH11010) - Bug when constructing
DataFramewhere passing a dictionary with only scalar values and specifying columns did not raise an error (GH10856) - Bug in
.var()causing roundoff errors for highly similar values (GH10242) - Bug in
DataFrame.plot(subplots=True)with duplicated columns outputs incorrect result (GH10962) - Bug in
Indexarithmetic may result in incorrect class (GH10638) - Bug in
date_rangeresults in empty if freq is negative annualy, quarterly and monthly (GH11018) - Bug in
DatetimeIndexcannot infer negative freq (GH11018) - Remove use of some deprecated numpy comparison operations, mainly in tests. (GH10569)
- Bug in
Indexdtype may not applied properly (GH11017) - Bug in
io.gbqwhen testing for minimum google api client version (GH10652) - Bug in
DataFrameconstruction from nesteddictwithtimedeltakeys (GH11129) - Bug in
.fillnaagainst may raiseTypeErrorwhen data contains datetime dtype (GH7095, GH11153) - Bug in
.groupbywhen number of keys to group by is same as length of index (GH11185) - Bug in
convert_objectswhere converted values might not be returned if all null andcoerce(GH9589) - Bug in
convert_objectswherecopykeyword was not respected (GH9589)
v0.16.2 (June 12, 2015)¶
This is a minor bug-fix release from 0.16.1 and includes a a large number of
bug fixes along some new features (pipe() method), enhancements, and performance improvements.
We recommend that all users upgrade to this version.
Highlights include:
What’s new in v0.16.2
New features¶
Pipe¶
We’ve introduced a new method DataFrame.pipe(). As suggested by the name, pipe
should be used to pipe data through a chain of function calls.
The goal is to avoid confusing nested function calls like
# df is a DataFrame
# f, g, and h are functions that take and return DataFrames
f(g(h(df), arg1=1), arg2=2, arg3=3)
The logic flows from inside out, and function names are separated from their keyword arguments. This can be rewritten as
(df.pipe(h)
.pipe(g, arg1=1)
.pipe(f, arg2=2, arg3=3)
)
Now both the code and the logic flow from top to bottom. Keyword arguments are next to their functions. Overall the code is much more readable.
In the example above, the functions f, g, and h each expected the DataFrame as the first positional argument.
When the function you wish to apply takes its data anywhere other than the first argument, pass a tuple
of (function, keyword) indicating where the DataFrame should flow. For example:
In [1]: import statsmodels.formula.api as sm
In [2]: bb = pd.read_csv('data/baseball.csv', index_col='id')
# sm.poisson takes (formula, data)
In [3]: (bb.query('h > 0')
...: .assign(ln_h = lambda df: np.log(df.h))
...: .pipe((sm.poisson, 'data'), 'hr ~ ln_h + year + g + C(lg)')
...: .fit()
...: .summary()
...: )
...:
Optimization terminated successfully.
Current function value: 2.116284
Iterations 24
Out[3]:
<class 'statsmodels.iolib.summary.Summary'>
"""
Poisson Regression Results
==============================================================================
Dep. Variable: hr No. Observations: 68
Model: Poisson Df Residuals: 63
Method: MLE Df Model: 4
Date: Fri, 07 Jul 2017 Pseudo R-squ.: 0.6878
Time: 12:29:29 Log-Likelihood: -143.91
converged: True LL-Null: -460.91
LLR p-value: 6.774e-136
===============================================================================
coef std err z P>|z| [0.025 0.975]
-------------------------------------------------------------------------------
Intercept -1267.3636 457.867 -2.768 0.006 -2164.767 -369.960
C(lg)[T.NL] -0.2057 0.101 -2.044 0.041 -0.403 -0.008
ln_h 0.9280 0.191 4.866 0.000 0.554 1.302
year 0.6301 0.228 2.762 0.006 0.183 1.077
g 0.0099 0.004 2.754 0.006 0.003 0.017
===============================================================================
"""
The pipe method is inspired by unix pipes, which stream text through
processes. More recently dplyr and magrittr have introduced the
popular (%>%) pipe operator for R.
See the documentation for more. (GH10129)
Other Enhancements¶
Added rsplit to Index/Series StringMethods (GH10303)
Removed the hard-coded size limits on the
DataFrameHTML representation in the IPython notebook, and leave this to IPython itself (only for IPython v3.0 or greater). This eliminates the duplicate scroll bars that appeared in the notebook with large frames (GH10231).Note that the notebook has a
toggle output scrollingfeature to limit the display of very large frames (by clicking left of the output). You can also configure the way DataFrames are displayed using the pandas options, see here here.axisparameter ofDataFrame.quantilenow accepts alsoindexandcolumn. (GH9543)
API Changes¶
Holidaynow raisesNotImplementedErrorif bothoffsetandobservanceare used in the constructor instead of returning an incorrect result (GH10217).
Performance Improvements¶
Bug Fixes¶
- Bug in
Series.histraises an error when a one rowSerieswas given (GH10214) - Bug where
HDFStore.selectmodifies the passed columns list (GH7212) - Bug in
Categoricalrepr withdisplay.widthofNonein Python 3 (GH10087) - Bug in
to_jsonwith certain orients and aCategoricalIndexwould segfault (GH10317) - Bug where some of the nan funcs do not have consistent return dtypes (GH10251)
- Bug in
DataFrame.quantileon checking that a valid axis was passed (GH9543) - Bug in
groupby.applyaggregation forCategoricalnot preserving categories (GH10138) - Bug in
to_csvwheredate_formatis ignored if thedatetimeis fractional (GH10209) - Bug in
DataFrame.to_jsonwith mixed data types (GH10289) - Bug in cache updating when consolidating (GH10264)
- Bug in
mean()where integer dtypes can overflow (GH10172) - Bug where
Panel.from_dictdoes not set dtype when specified (GH10058) - Bug in
Index.unionraisesAttributeErrorwhen passing array-likes. (GH10149) - Bug in
Timestamp‘s’microsecond,quarter,dayofyear,weekanddaysinmonthproperties returnnp.inttype, not built-inint. (GH10050) - Bug in
NaTraisesAttributeErrorwhen accessing todaysinmonth,dayofweekproperties. (GH10096) - Bug in Index repr when using the
max_seq_items=Nonesetting (GH10182). - Bug in getting timezone data with
dateutilon various platforms ( GH9059, GH8639, GH9663, GH10121) - Bug in displaying datetimes with mixed frequencies; display ‘ms’ datetimes to the proper precision. (GH10170)
- Bug in
setitemwhere type promotion is applied to the entire block (GH10280) - Bug in
Seriesarithmetic methods may incorrectly hold names (GH10068) - Bug in
GroupBy.get_groupwhen grouping on multiple keys, one of which is categorical. (GH10132) - Bug in
DatetimeIndexandTimedeltaIndexnames are lost after timedelta arithmetics ( GH9926) - Bug in
DataFrameconstruction from nesteddictwithdatetime64(GH10160) - Bug in
Seriesconstruction fromdictwithdatetime64keys (GH9456) - Bug in
Series.plot(label="LABEL")not correctly setting the label (GH10119) - Bug in
plotnot defaulting to matplotlibaxes.gridsetting (GH9792) - Bug causing strings containing an exponent, but no decimal to be parsed as
intinstead offloatinengine='python'for theread_csvparser (GH9565) - Bug in
Series.alignresetsnamewhenfill_valueis specified (GH10067) - Bug in
read_csvcausing index name not to be set on an empty DataFrame (GH10184) - Bug in
SparseSeries.absresetsname(GH10241) - Bug in
TimedeltaIndexslicing may reset freq (GH10292) - Bug in
GroupBy.get_groupraisesValueErrorwhen group key containsNaT(GH6992) - Bug in
SparseSeriesconstructor ignores input data name (GH10258) - Bug in
Categorical.remove_categoriescausing aValueErrorwhen removing theNaNcategory if underlying dtype is floating-point (GH10156) - Bug where infer_freq infers timerule (WOM-5XXX) unsupported by to_offset (GH9425)
- Bug in
DataFrame.to_hdf()where table format would raise a seemingly unrelated error for invalid (non-string) column names. This is now explicitly forbidden. (GH9057) - Bug to handle masking empty
DataFrame(GH10126). - Bug where MySQL interface could not handle numeric table/column names (GH10255)
- Bug in
read_csvwith adate_parserthat returned adatetime64array of other time resolution than[ns](GH10245) - Bug in
Panel.applywhen the result has ndim=0 (GH10332) - Bug in
read_hdfwhereauto_closecould not be passed (GH9327). - Bug in
read_hdfwhere open stores could not be used (GH10330). - Bug in adding empty
DataFrame``s, now results in a ``DataFramethat.equalsan emptyDataFrame(GH10181). - Bug in
to_hdfandHDFStorewhich did not check that complib choices were valid (GH4582, GH8874).
v0.16.1 (May 11, 2015)¶
This is a minor bug-fix release from 0.16.0 and includes a a large number of bug fixes along several new features, enhancements, and performance improvements. We recommend that all users upgrade to this version.
Highlights include:
- Support for a
CategoricalIndex, a category based index, see here - New section on how-to-contribute to pandas, see here
- Revised “Merge, join, and concatenate” documentation, including graphical examples to make it easier to understand each operations, see here
- New method
samplefor drawing random samples from Series, DataFrames and Panels. See here - The default
Indexprinting has changed to a more uniform format, see here BusinessHourdatetime-offset is now supported, see here- Further enhancement to the
.straccessor to make string operations easier, see here
What’s new in v0.16.1
Warning
In pandas 0.17.0, the sub-package pandas.io.data will be removed in favor of a separately installable package. See here for details (GH8961)
Enhancements¶
CategoricalIndex¶
We introduce a CategoricalIndex, a new type of index object that is useful for supporting
indexing with duplicates. This is a container around a Categorical (introduced in v0.15.0)
and allows efficient indexing and storage of an index with a large number of duplicated elements. Prior to 0.16.1,
setting the index of a DataFrame/Series with a category dtype would convert this to regular object-based Index.
In [1]: df = DataFrame({'A' : np.arange(6),
...: 'B' : Series(list('aabbca')).astype('category',
...: categories=list('cab'))
...: })
...:
In [2]: df
Out[2]:
A B
0 0 a
1 1 a
2 2 b
3 3 b
4 4 c
5 5 a
In [3]: df.dtypes
�����������������������������������������������������������������Out[3]:
A int64
B category
dtype: object
In [4]: df.B.cat.categories
��������������������������������������������������������������������������������������������������������������������Out[4]: Index(['c', 'a', 'b'], dtype='object')
setting the index, will create create a CategoricalIndex
In [5]: df2 = df.set_index('B')
In [6]: df2.index
Out[6]: CategoricalIndex(['a', 'a', 'b', 'b', 'c', 'a'], categories=['c', 'a', 'b'], ordered=False, name='B', dtype='category')
indexing with __getitem__/.iloc/.loc/.ix works similarly to an Index with duplicates.
The indexers MUST be in the category or the operation will raise.
In [7]: df2.loc['a']
Out[7]:
A
B
a 0
a 1
a 5
and preserves the CategoricalIndex
In [8]: df2.loc['a'].index
Out[8]: CategoricalIndex(['a', 'a', 'a'], categories=['c', 'a', 'b'], ordered=False, name='B', dtype='category')
sorting will order by the order of the categories
In [9]: df2.sort_index()
Out[9]:
A
B
c 4
a 0
a 1
a 5
b 2
b 3
groupby operations on the index will preserve the index nature as well
In [10]: df2.groupby(level=0).sum()
Out[10]:
A
B
c 4
a 6
b 5
In [11]: df2.groupby(level=0).sum().index
�����������������������������������Out[11]: CategoricalIndex(['c', 'a', 'b'], categories=['c', 'a', 'b'], ordered=False, name='B', dtype='category')
reindexing operations, will return a resulting index based on the type of the passed
indexer, meaning that passing a list will return a plain-old-Index; indexing with
a Categorical will return a CategoricalIndex, indexed according to the categories
of the PASSED Categorical dtype. This allows one to arbitrarly index these even with
values NOT in the categories, similarly to how you can reindex ANY pandas index.
In [12]: df2.reindex(['a','e'])
Out[12]:
A
B
a 0.0
a 1.0
a 5.0
e NaN
In [13]: df2.reindex(['a','e']).index
����������������������������������������������������Out[13]: Index(['a', 'a', 'a', 'e'], dtype='object', name='B')
In [14]: df2.reindex(pd.Categorical(['a','e'],categories=list('abcde')))
�������������������������������������������������������������������������������������������������������������������Out[14]:
A
B
a 0.0
a 1.0
a 5.0
e NaN
In [15]: df2.reindex(pd.Categorical(['a','e'],categories=list('abcde'))).index
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[15]: CategoricalIndex(['a', 'a', 'a', 'e'], categories=['a', 'b', 'c', 'd', 'e'], ordered=False, name='B', dtype='category')
See the documentation for more. (GH7629, GH10038, GH10039)
Sample¶
Series, DataFrames, and Panels now have a new method: sample().
The method accepts a specific number of rows or columns to return, or a fraction of the
total number or rows or columns. It also has options for sampling with or without replacement,
for passing in a column for weights for non-uniform sampling, and for setting seed values to
facilitate replication. (GH2419)
In [16]: example_series = Series([0,1,2,3,4,5])
# When no arguments are passed, returns 1
In [17]: example_series.sample()
Out[17]:
3 3
dtype: int64
# One may specify either a number of rows:
In [18]: example_series.sample(n=3)
������������������������������Out[18]:
5 5
1 1
4 4
dtype: int64
# Or a fraction of the rows:
In [19]: example_series.sample(frac=0.5)
��������������������������������������������������������������������������Out[19]:
4 4
1 1
0 0
dtype: int64
# weights are accepted.
In [20]: example_weights = [0, 0, 0.2, 0.2, 0.2, 0.4]
In [21]: example_series.sample(n=3, weights=example_weights)
Out[21]:
2 2
3 3
5 5
dtype: int64
# weights will also be normalized if they do not sum to one,
# and missing values will be treated as zeros.
In [22]: example_weights2 = [0.5, 0, 0, 0, None, np.nan]
In [23]: example_series.sample(n=1, weights=example_weights2)
Out[23]:
0 0
dtype: int64
When applied to a DataFrame, one may pass the name of a column to specify sampling weights when sampling from rows.
In [24]: df = DataFrame({'col1':[9,8,7,6], 'weight_column':[0.5, 0.4, 0.1, 0]})
In [25]: df.sample(n=3, weights='weight_column')
Out[25]:
col1 weight_column
0 9 0.5
1 8 0.4
2 7 0.1
String Methods Enhancements¶
Continuing from v0.16.0, the following enhancements make string operations easier and more consistent with standard python string operations.
Added
StringMethods(.straccessor) toIndex(GH9068)The
.straccessor is now available for bothSeriesandIndex.In [26]: idx = Index([' jack', 'jill ', ' jesse ', 'frank']) In [27]: idx.str.strip() Out[27]: Index(['jack', 'jill', 'jesse', 'frank'], dtype='object')
One special case for the .str accessor on
Indexis that if a string method returnsbool, the.straccessor will return anp.arrayinstead of a booleanIndex(GH8875). This enables the following expression to work naturally:In [28]: idx = Index(['a1', 'a2', 'b1', 'b2']) In [29]: s = Series(range(4), index=idx) In [30]: s Out[30]: a1 0 a2 1 b1 2 b2 3 dtype: int64 In [31]: idx.str.startswith('a') �������������������������������������������������������Out[31]: array([ True, True, False, False], dtype=bool) In [32]: s[s.index.str.startswith('a')] ����������������������������������������������������������������������������������������������������������������Out[32]: a1 0 a2 1 dtype: int64
The following new methods are accesible via
.straccessor to apply the function to each values. (GH9766, GH9773, GH10031, GH10045, GH10052)Methods capitalize()swapcase()normalize()partition()rpartition()index()rindex()translate()splitnow takesexpandkeyword to specify whether to expand dimensionality.return_typeis deprecated. (GH9847)In [33]: s = Series(['a,b', 'a,c', 'b,c']) # return Series In [34]: s.str.split(',') Out[34]: 0 [a, b] 1 [a, c] 2 [b, c] dtype: object # return DataFrame In [35]: s.str.split(',', expand=True) ������������������������������������������������������������Out[35]: 0 1 0 a b 1 a c 2 b c In [36]: idx = Index(['a,b', 'a,c', 'b,c']) # return Index In [37]: idx.str.split(',') Out[37]: Index([['a', 'b'], ['a', 'c'], ['b', 'c']], dtype='object') # return MultiIndex In [38]: idx.str.split(',', expand=True) ���������������������������������������������������������������������Out[38]: MultiIndex(levels=[['a', 'b'], ['b', 'c']], labels=[[0, 0, 1], [0, 1, 1]])
Improved
extractandget_dummiesmethods forIndex.str(GH9980)
Other Enhancements¶
BusinessHouroffset is now supported, which represents business hours starting from 09:00 - 17:00 onBusinessDayby default. See Here for details. (GH7905)In [39]: from pandas.tseries.offsets import BusinessHour In [40]: Timestamp('2014-08-01 09:00') + BusinessHour() Out[40]: Timestamp('2014-08-01 10:00:00') In [41]: Timestamp('2014-08-01 07:00') + BusinessHour() ������������������������������������������Out[41]: Timestamp('2014-08-01 10:00:00') In [42]: Timestamp('2014-08-01 16:30') + BusinessHour() ������������������������������������������������������������������������������������Out[42]: Timestamp('2014-08-04 09:30:00')
DataFrame.diffnow takes anaxisparameter that determines the direction of differencing (GH9727)Allow
clip,clip_lower, andclip_upperto accept array-like arguments as thresholds (This is a regression from 0.11.0). These methods now have anaxisparameter which determines how the Series or DataFrame will be aligned with the threshold(s). (GH6966)DataFrame.mask()andSeries.mask()now support same keywords aswhere(GH8801)dropfunction can now accepterrorskeyword to suppressValueErrorraised when any of label does not exist in the target data. (GH6736)In [43]: df = DataFrame(np.random.randn(3, 3), columns=['A', 'B', 'C']) In [44]: df.drop(['A', 'X'], axis=1, errors='ignore') Out[44]: B C 0 1.058969 -0.397840 1 1.047579 1.045938 2 -0.122092 0.124713
Add support for separating years and quarters using dashes, for example 2014-Q1. (GH9688)
Allow conversion of values with dtype
datetime64ortimedelta64to strings usingastype(str)(GH9757)get_dummiesfunction now acceptssparsekeyword. If set toTrue, the returnDataFrameis sparse, e.g.SparseDataFrame. (GH8823)Periodnow acceptsdatetime64as value input. (GH9054)Allow timedelta string conversion when leading zero is missing from time definition, ie 0:00:00 vs 00:00:00. (GH9570)
Allow
Panel.shiftwithaxis='items'(GH9890)Trying to write an excel file now raises
NotImplementedErrorif theDataFramehas aMultiIndexinstead of writing a broken Excel file. (GH9794)Allow
Categorical.add_categoriesto acceptSeriesornp.array. (GH9927)Add/delete
str/dt/cataccessors dynamically from__dir__. (GH9910)Add
normalizeas adtaccessor method. (GH10047)DataFrameandSeriesnow have_constructor_expanddimproperty as overridable constructor for one higher dimensionality data. This should be used only when it is really needed, see herepd.lib.infer_dtypenow returns'bytes'in Python 3 where appropriate. (GH10032)
API changes¶
- When passing in an ax to
df.plot( ..., ax=ax), the sharex kwarg will now default to False. The result is that the visibility of xlabels and xticklabels will not anymore be changed. You have to do that by yourself for the right axes in your figure or setsharex=Trueexplicitly (but this changes the visible for all axes in the figure, not only the one which is passed in!). If pandas creates the subplots itself (e.g. no passed in ax kwarg), then the default is stillsharex=Trueand the visibility changes are applied. assign()now inserts new columns in alphabetical order. Previously the order was arbitrary. (GH9777)- By default,
read_csvandread_tablewill now try to infer the compression type based on the file extension. Setcompression=Noneto restore the previous behavior (no decompression). (GH9770)
Index Representation¶
The string representation of Index and its sub-classes have now been unified. These will show a single-line display if there are few values; a wrapped multi-line display for a lot of values (but less than display.max_seq_items; if lots of items (> display.max_seq_items) will show a truncated display (the head and tail of the data). The formatting for MultiIndex is unchanges (a multi-line wrapped display). The display width responds to the option display.max_seq_items, which is defaulted to 100. (GH6482)
Previous Behavior
In [2]: pd.Index(range(4),name='foo')
Out[2]: Int64Index([0, 1, 2, 3], dtype='int64')
In [3]: pd.Index(range(104),name='foo')
Out[3]: Int64Index([0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99, ...], dtype='int64')
In [4]: pd.date_range('20130101',periods=4,name='foo',tz='US/Eastern')
Out[4]:
<class 'pandas.tseries.index.DatetimeIndex'>
[2013-01-01 00:00:00-05:00, ..., 2013-01-04 00:00:00-05:00]
Length: 4, Freq: D, Timezone: US/Eastern
In [5]: pd.date_range('20130101',periods=104,name='foo',tz='US/Eastern')
Out[5]:
<class 'pandas.tseries.index.DatetimeIndex'>
[2013-01-01 00:00:00-05:00, ..., 2013-04-14 00:00:00-04:00]
Length: 104, Freq: D, Timezone: US/Eastern
New Behavior
In [45]: pd.set_option('display.width', 80)
In [46]: pd.Index(range(4), name='foo')
Out[46]: RangeIndex(start=0, stop=4, step=1, name='foo')
In [47]: pd.Index(range(30), name='foo')
���������������������������������������������������������Out[47]: RangeIndex(start=0, stop=30, step=1, name='foo')
In [48]: pd.Index(range(104), name='foo')
�������������������������������������������������������������������������������������������������������������������Out[48]: RangeIndex(start=0, stop=104, step=1, name='foo')
In [49]: pd.CategoricalIndex(['a','bb','ccc','dddd'], ordered=True, name='foobar')
������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[49]: CategoricalIndex(['a', 'bb', 'ccc', 'dddd'], categories=['a', 'bb', 'ccc', 'dddd'], ordered=True, name='foobar', dtype='category')
In [50]: pd.CategoricalIndex(['a','bb','ccc','dddd']*10, ordered=True, name='foobar')
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[50]:
CategoricalIndex(['a', 'bb', 'ccc', 'dddd', 'a', 'bb', 'ccc', 'dddd', 'a',
'bb', 'ccc', 'dddd', 'a', 'bb', 'ccc', 'dddd', 'a', 'bb',
'ccc', 'dddd', 'a', 'bb', 'ccc', 'dddd', 'a', 'bb', 'ccc',
'dddd', 'a', 'bb', 'ccc', 'dddd', 'a', 'bb', 'ccc', 'dddd',
'a', 'bb', 'ccc', 'dddd'],
categories=['a', 'bb', 'ccc', 'dddd'], ordered=True, name='foobar', dtype='category')
In [51]: pd.CategoricalIndex(['a','bb','ccc','dddd']*100, ordered=True, name='foobar')
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[51]:
CategoricalIndex(['a', 'bb', 'ccc', 'dddd', 'a', 'bb', 'ccc', 'dddd', 'a',
'bb',
...
'ccc', 'dddd', 'a', 'bb', 'ccc', 'dddd', 'a', 'bb', 'ccc',
'dddd'],
categories=['a', 'bb', 'ccc', 'dddd'], ordered=True, name='foobar', dtype='category', length=400)
In [52]: pd.date_range('20130101',periods=4, name='foo', tz='US/Eastern')
������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[52]:
DatetimeIndex(['2013-01-01 00:00:00-05:00', '2013-01-02 00:00:00-05:00',
'2013-01-03 00:00:00-05:00', '2013-01-04 00:00:00-05:00'],
dtype='datetime64[ns, US/Eastern]', name='foo', freq='D')
In [53]: pd.date_range('20130101',periods=25, freq='D')
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[53]:
DatetimeIndex(['2013-01-01', '2013-01-02', '2013-01-03', '2013-01-04',
'2013-01-05', '2013-01-06', '2013-01-07', '2013-01-08',
'2013-01-09', '2013-01-10', '2013-01-11', '2013-01-12',
'2013-01-13', '2013-01-14', '2013-01-15', '2013-01-16',
'2013-01-17', '2013-01-18', '2013-01-19', '2013-01-20',
'2013-01-21', '2013-01-22', '2013-01-23', '2013-01-24',
'2013-01-25'],
dtype='datetime64[ns]', freq='D')
In [54]: pd.date_range('20130101',periods=104, name='foo', tz='US/Eastern')
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[54]:
DatetimeIndex(['2013-01-01 00:00:00-05:00', '2013-01-02 00:00:00-05:00',
'2013-01-03 00:00:00-05:00', '2013-01-04 00:00:00-05:00',
'2013-01-05 00:00:00-05:00', '2013-01-06 00:00:00-05:00',
'2013-01-07 00:00:00-05:00', '2013-01-08 00:00:00-05:00',
'2013-01-09 00:00:00-05:00', '2013-01-10 00:00:00-05:00',
...
'2013-04-05 00:00:00-04:00', '2013-04-06 00:00:00-04:00',
'2013-04-07 00:00:00-04:00', '2013-04-08 00:00:00-04:00',
'2013-04-09 00:00:00-04:00', '2013-04-10 00:00:00-04:00',
'2013-04-11 00:00:00-04:00', '2013-04-12 00:00:00-04:00',
'2013-04-13 00:00:00-04:00', '2013-04-14 00:00:00-04:00'],
dtype='datetime64[ns, US/Eastern]', name='foo', length=104, freq='D')
Performance Improvements¶
Bug Fixes¶
- Bug where labels did not appear properly in the legend of
DataFrame.plot(), passinglabel=arguments works, and Series indices are no longer mutated. (GH9542) - Bug in json serialization causing a segfault when a frame had zero length. (GH9805)
- Bug in
read_csvwhere missing trailing delimiters would cause segfault. (GH5664) - Bug in retaining index name on appending (GH9862)
- Bug in
scatter_matrixdraws unexpected axis ticklabels (GH5662) - Fixed bug in
StataWriterresulting in changes to inputDataFrameupon save (GH9795). - Bug in
transformcausing length mismatch when null entries were present and a fast aggregator was being used (GH9697) - Bug in
equalscausing false negatives when block order differed (GH9330) - Bug in grouping with multiple
pd.Grouperwhere one is non-time based (GH10063) - Bug in
read_sql_tableerror when reading postgres table with timezone (GH7139) - Bug in
DataFrameslicing may not retain metadata (GH9776) - Bug where
TimdeltaIndexwere not properly serialized in fixedHDFStore(GH9635) - Bug with
TimedeltaIndexconstructor ignoringnamewhen given anotherTimedeltaIndexas data (GH10025). - Bug in
DataFrameFormatter._get_formatted_indexwith not applyingmax_colwidthto theDataFrameindex (GH7856) - Bug in
.locwith a read-only ndarray data source (GH10043) - Bug in
groupby.apply()that would raise if a passed user defined function either returned onlyNone(for all input). (GH9685) - Always use temporary files in pytables tests (GH9992)
- Bug in plotting continuously using
secondary_ymay not show legend properly. (GH9610, GH9779) - Bug in
DataFrame.plot(kind="hist")results inTypeErrorwhenDataFramecontains non-numeric columns (GH9853) - Bug where repeated plotting of
DataFramewith aDatetimeIndexmay raiseTypeError(GH9852) - Bug in
setup.pythat would allow an incompat cython version to build (GH9827) - Bug in plotting
secondary_yincorrectly attachesright_axproperty to secondary axes specifying itself recursively. (GH9861) - Bug in
Series.quantileon empty Series of typeDatetimeorTimedelta(GH9675) - Bug in
wherecausing incorrect results when upcasting was required (GH9731) - Bug in
FloatArrayFormatterwhere decision boundary for displaying “small” floats in decimal format is off by one order of magnitude for a given display.precision (GH9764) - Fixed bug where
DataFrame.plot()raised an error when bothcolorandstylekeywords were passed and there was no color symbol in the style strings (GH9671) - Not showing a
DeprecationWarningon combining list-likes with anIndex(GH10083) - Bug in
read_csvandread_tablewhen usingskip_rowsparameter if blank lines are present. (GH9832) - Bug in
read_csv()interpretsindex_col=Trueas1(GH9798) - Bug in index equality comparisons using
==failing on Index/MultiIndex type incompatibility (GH9785) - Bug in which
SparseDataFramecould not take nan as a column name (GH8822) - Bug in
to_msgpackandread_msgpackzlib and blosc compression support (GH9783) - Bug
GroupBy.sizedoesn’t attach index name properly if grouped byTimeGrouper(GH9925) - Bug causing an exception in slice assignments because
length_of_indexerreturns wrong results (GH9995) - Bug in csv parser causing lines with initial whitespace plus one non-space character to be skipped. (GH9710)
- Bug in C csv parser causing spurious NaNs when data started with newline followed by whitespace. (GH10022)
- Bug causing elements with a null group to spill into the final group when grouping by a
Categorical(GH9603) - Bug where .iloc and .loc behavior is not consistent on empty dataframes (GH9964)
- Bug in invalid attribute access on a
TimedeltaIndexincorrectly raisedValueErrorinstead ofAttributeError(GH9680) - Bug in unequal comparisons between categorical data and a scalar, which was not in the categories (e.g.
Series(Categorical(list("abc"), ordered=True)) > "d". This returnedFalsefor all elements, but now raises aTypeError. Equality comparisons also now returnFalsefor==andTruefor!=. (GH9848) - Bug in DataFrame
__setitem__when right hand side is a dictionary (GH9874) - Bug in
wherewhen dtype isdatetime64/timedelta64, but dtype of other is not (GH9804) - Bug in
MultiIndex.sortlevel()results in unicode level name breaks (GH9856) - Bug in which
groupby.transformincorrectly enforced output dtypes to match input dtypes. (GH9807) - Bug in
DataFrameconstructor whencolumnsparameter is set, anddatais an empty list (GH9939) - Bug in bar plot with
log=TrueraisesTypeErrorif all values are less than 1 (GH9905) - Bug in horizontal bar plot ignores
log=True(GH9905) - Bug in PyTables queries that did not return proper results using the index (GH8265, GH9676)
- Bug where dividing a dataframe containing values of type
Decimalby anotherDecimalwould raise. (GH9787) - Bug where using DataFrames asfreq would remove the name of the index. (GH9885)
- Bug causing extra index point when resample BM/BQ (GH9756)
- Changed caching in
AbstractHolidayCalendarto be at the instance level rather than at the class level as the latter can result in unexpected behaviour. (GH9552) - Fixed latex output for multi-indexed dataframes (GH9778)
- Bug causing an exception when setting an empty range using
DataFrame.loc(GH9596) - Bug in hiding ticklabels with subplots and shared axes when adding a new plot to an existing grid of axes (GH9158)
- Bug in
transformandfilterwhen grouping on a categorical variable (GH9921) - Bug in
transformwhen groups are equal in number and dtype to the input index (GH9700) - Google BigQuery connector now imports dependencies on a per-method basis.(GH9713)
- Updated BigQuery connector to no longer use deprecated
oauth2client.tools.run()(GH8327) - Bug in subclassed
DataFrame. It may not return the correct class, when slicing or subsetting it. (GH9632) - Bug in
.median()where non-float null values are not handled correctly (GH10040) - Bug in Series.fillna() where it raises if a numerically convertible string is given (GH10092)
v0.16.0 (March 22, 2015)¶
This is a major release from 0.15.2 and includes a small number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
Highlights include:
DataFrame.assignmethod, see hereSeries.to_coo/from_coomethods to interact withscipy.sparse, see here- Backwards incompatible change to
Timedeltato conform the.secondsattribute withdatetime.timedelta, see here - Changes to the
.locslicing API to conform with the behavior of.ixsee here - Changes to the default for ordering in the
Categoricalconstructor, see here - Enhancement to the
.straccessor to make string operations easier, see here - The
pandas.tools.rplot,pandas.sandbox.qtpandasandpandas.rpymodules are deprecated. We refer users to external packages like seaborn, pandas-qt and rpy2 for similar or equivalent functionality, see here
Check the API Changes and deprecations before updating.
What’s new in v0.16.0
New features¶
DataFrame Assign¶
Inspired by dplyr’s mutate verb, DataFrame has a new
assign() method.
The function signature for assign is simply **kwargs. The keys
are the column names for the new fields, and the values are either a value
to be inserted (for example, a Series or NumPy array), or a function
of one argument to be called on the DataFrame. The new values are inserted,
and the entire DataFrame (with all original and new columns) is returned.
In [1]: iris = read_csv('data/iris.data')
In [2]: iris.head()
Out[2]:
SepalLength SepalWidth PetalLength PetalWidth Name
0 5.1 3.5 1.4 0.2 Iris-setosa
1 4.9 3.0 1.4 0.2 Iris-setosa
2 4.7 3.2 1.3 0.2 Iris-setosa
3 4.6 3.1 1.5 0.2 Iris-setosa
4 5.0 3.6 1.4 0.2 Iris-setosa
In [3]: iris.assign(sepal_ratio=iris['SepalWidth'] / iris['SepalLength']).head()
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[3]:
SepalLength SepalWidth PetalLength PetalWidth Name sepal_ratio
0 5.1 3.5 1.4 0.2 Iris-setosa 0.686275
1 4.9 3.0 1.4 0.2 Iris-setosa 0.612245
2 4.7 3.2 1.3 0.2 Iris-setosa 0.680851
3 4.6 3.1 1.5 0.2 Iris-setosa 0.673913
4 5.0 3.6 1.4 0.2 Iris-setosa 0.720000
Above was an example of inserting a precomputed value. We can also pass in a function to be evalutated.
In [4]: iris.assign(sepal_ratio = lambda x: (x['SepalWidth'] /
...: x['SepalLength'])).head()
...:
Out[4]:
SepalLength SepalWidth PetalLength PetalWidth Name sepal_ratio
0 5.1 3.5 1.4 0.2 Iris-setosa 0.686275
1 4.9 3.0 1.4 0.2 Iris-setosa 0.612245
2 4.7 3.2 1.3 0.2 Iris-setosa 0.680851
3 4.6 3.1 1.5 0.2 Iris-setosa 0.673913
4 5.0 3.6 1.4 0.2 Iris-setosa 0.720000
The power of assign comes when used in chains of operations. For example,
we can limit the DataFrame to just those with a Sepal Length greater than 5,
calculate the ratio, and plot
In [5]: (iris.query('SepalLength > 5')
...: .assign(SepalRatio = lambda x: x.SepalWidth / x.SepalLength,
...: PetalRatio = lambda x: x.PetalWidth / x.PetalLength)
...: .plot(kind='scatter', x='SepalRatio', y='PetalRatio'))
...:
Out[5]: <matplotlib.axes._subplots.AxesSubplot at 0x13df38208>
See the documentation for more. (GH9229)
Interaction with scipy.sparse¶
Added SparseSeries.to_coo() and SparseSeries.from_coo() methods (GH8048) for converting to and from scipy.sparse.coo_matrix instances (see here). For example, given a SparseSeries with MultiIndex we can convert to a scipy.sparse.coo_matrix by specifying the row and column labels as index levels:
In [6]: from numpy import nan
In [7]: s = Series([3.0, nan, 1.0, 3.0, nan, nan])
In [8]: s.index = MultiIndex.from_tuples([(1, 2, 'a', 0),
...: (1, 2, 'a', 1),
...: (1, 1, 'b', 0),
...: (1, 1, 'b', 1),
...: (2, 1, 'b', 0),
...: (2, 1, 'b', 1)],
...: names=['A', 'B', 'C', 'D'])
...:
In [9]: s
Out[9]:
A B C D
1 2 a 0 3.0
1 NaN
1 b 0 1.0
1 3.0
2 1 b 0 NaN
1 NaN
dtype: float64
# SparseSeries
In [10]: ss = s.to_sparse()
In [11]: ss
Out[11]:
A B C D
1 2 a 0 3.0
1 NaN
1 b 0 1.0
1 3.0
2 1 b 0 NaN
1 NaN
dtype: float64
BlockIndex
Block locations: array([0, 2], dtype=int32)
Block lengths: array([1, 2], dtype=int32)
In [12]: A, rows, columns = ss.to_coo(row_levels=['A', 'B'],
....: column_levels=['C', 'D'],
....: sort_labels=False)
....:
In [13]: A
Out[13]:
<3x4 sparse matrix of type '<class 'numpy.float64'>'
with 3 stored elements in COOrdinate format>
In [14]: A.todense()
�������������������������������������������������������������������������������������������������������������Out[14]:
matrix([[ 3., 0., 0., 0.],
[ 0., 0., 1., 3.],
[ 0., 0., 0., 0.]])
In [15]: rows
������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[15]: [(1, 2), (1, 1), (2, 1)]
In [16]: columns
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[16]: [('a', 0), ('a', 1), ('b', 0), ('b', 1)]
The from_coo method is a convenience method for creating a SparseSeries
from a scipy.sparse.coo_matrix:
In [17]: from scipy import sparse
In [18]: A = sparse.coo_matrix(([3.0, 1.0, 2.0], ([1, 0, 0], [0, 2, 3])),
....: shape=(3, 4))
....:
In [19]: A
Out[19]:
<3x4 sparse matrix of type '<class 'numpy.float64'>'
with 3 stored elements in COOrdinate format>
In [20]: A.todense()
�������������������������������������������������������������������������������������������������������������Out[20]:
matrix([[ 0., 0., 1., 2.],
[ 3., 0., 0., 0.],
[ 0., 0., 0., 0.]])
In [21]: ss = SparseSeries.from_coo(A)
In [22]: ss
Out[22]:
0 2 1.0
3 2.0
1 0 3.0
dtype: float64
BlockIndex
Block locations: array([0], dtype=int32)
Block lengths: array([3], dtype=int32)
String Methods Enhancements¶
Following new methods are accesible via
.straccessor to apply the function to each values. This is intended to make it more consistent with standard methods on strings. (GH9282, GH9352, GH9386, GH9387, GH9439)Methods isalnum()isalpha()isdigit()isdigit()isspace()islower()isupper()istitle()isnumeric()isdecimal()find()rfind()ljust()rjust()zfill()In [23]: s = Series(['abcd', '3456', 'EFGH']) In [24]: s.str.isalpha() Out[24]: 0 True 1 False 2 True dtype: bool In [25]: s.str.find('ab') �������������������������������������������������������Out[25]: 0 0 1 -1 2 -1 dtype: int64
Series.str.pad()andSeries.str.center()now acceptfillcharoption to specify filling character (GH9352)In [26]: s = Series(['12', '300', '25']) In [27]: s.str.pad(5, fillchar='_') Out[27]: 0 ___12 1 __300 2 ___25 dtype: object
Added
Series.str.slice_replace(), which previously raisedNotImplementedError(GH8888)In [28]: s = Series(['ABCD', 'EFGH', 'IJK']) In [29]: s.str.slice_replace(1, 3, 'X') Out[29]: 0 AXD 1 EXH 2 IX dtype: object # replaced with empty char In [30]: s.str.slice_replace(0, 1) ���������������������������������������������������Out[30]: 0 BCD 1 FGH 2 JK dtype: object
Other enhancements¶
Reindex now supports
method='nearest'for frames or series with a monotonic increasing or decreasing index (GH9258):In [31]: df = pd.DataFrame({'x': range(5)}) In [32]: df.reindex([0.2, 1.8, 3.5], method='nearest') Out[32]: x 0.2 0 1.8 2 3.5 4
This method is also exposed by the lower level
Index.get_indexerandIndex.get_locmethods.The
read_excel()function’s sheetname argument now accepts a list andNone, to get multiple or all sheets respectively. If more than one sheet is specified, a dictionary is returned. (GH9450)# Returns the 1st and 4th sheet, as a dictionary of DataFrames. pd.read_excel('path_to_file.xls',sheetname=['Sheet1',3])
Allow Stata files to be read incrementally with an iterator; support for long strings in Stata files. See the docs here (GH9493:).
Paths beginning with ~ will now be expanded to begin with the user’s home directory (GH9066)
Added time interval selection in
get_data_yahoo(GH9071)Added
Timestamp.to_datetime64()to complementTimedelta.to_timedelta64()(GH9255)tseries.frequencies.to_offset()now acceptsTimedeltaas input (GH9064)Lag parameter was added to the autocorrelation method of
Series, defaults to lag-1 autocorrelation (GH9192)Timedeltawill now acceptnanosecondskeyword in constructor (GH9273)SQL code now safely escapes table and column names (GH8986)
Added auto-complete for
Series.str.<tab>,Series.dt.<tab>andSeries.cat.<tab>(GH9322)Index.get_indexernow supportsmethod='pad'andmethod='backfill'even for any target array, not just monotonic targets. These methods also work for monotonic decreasing as well as monotonic increasing indexes (GH9258).Index.asofnow works on all index types (GH9258).A
verboseargument has been augmented inio.read_excel(), defaults to False. Set to True to print sheet names as they are parsed. (GH9450)Added
days_in_month(compatibility aliasdaysinmonth) property toTimestamp,DatetimeIndex,Period,PeriodIndex, andSeries.dt(GH9572)Added
decimaloption into_csvto provide formatting for non-‘.’ decimal separators (GH781)Added
normalizeoption forTimestampto normalized to midnight (GH8794)Added example for
DataFrameimport to R using HDF5 file andrhdf5library. See the documentation for more (GH9636).
Backwards incompatible API changes¶
Changes in Timedelta¶
In v0.15.0 a new scalar type Timedelta was introduced, that is a
sub-class of datetime.timedelta. Mentioned here was a notice of an API change w.r.t. the .seconds accessor. The intent was to provide a user-friendly set of accessors that give the ‘natural’ value for that unit, e.g. if you had a Timedelta('1 day, 10:11:12'), then .seconds would return 12. However, this is at odds with the definition of datetime.timedelta, which defines .seconds as 10 * 3600 + 11 * 60 + 12 == 36672.
So in v0.16.0, we are restoring the API to match that of datetime.timedelta. Further, the component values are still available through the .components accessor. This affects the .seconds and .microseconds accessors, and removes the .hours, .minutes, .milliseconds accessors. These changes affect TimedeltaIndex and the Series .dt accessor as well. (GH9185, GH9139)
Previous Behavior
In [2]: t = pd.Timedelta('1 day, 10:11:12.100123')
In [3]: t.days
Out[3]: 1
In [4]: t.seconds
Out[4]: 12
In [5]: t.microseconds
Out[5]: 123
New Behavior
In [33]: t = pd.Timedelta('1 day, 10:11:12.100123')
In [34]: t.days
Out[34]: 1
In [35]: t.seconds
�����������Out[35]: 36672
In [36]: t.microseconds
��������������������������Out[36]: 100123
Using .components allows the full component access
In [37]: t.components
Out[37]: Components(days=1, hours=10, minutes=11, seconds=12, milliseconds=100, microseconds=123, nanoseconds=0)
In [38]: t.components.seconds
�����������������������������������������������������������������������������������������������������������������Out[38]: 12
Indexing Changes¶
The behavior of a small sub-set of edge cases for using .loc have changed (GH8613). Furthermore we have improved the content of the error messages that are raised:
Slicing with
.locwhere the start and/or stop bound is not found in the index is now allowed; this previously would raise aKeyError. This makes the behavior the same as.ixin this case. This change is only for slicing, not when indexing with a single label.In [39]: df = DataFrame(np.random.randn(5,4), ....: columns=list('ABCD'), ....: index=date_range('20130101',periods=5)) ....: In [40]: df Out[40]: A B C D 2013-01-01 -0.322795 0.841675 2.390961 0.076200 2013-01-02 -0.566446 0.036142 -2.074978 0.247792 2013-01-03 -0.897157 -0.136795 0.018289 0.755414 2013-01-04 0.215269 0.841009 -1.445810 -1.401973 2013-01-05 -0.100918 -0.548242 -0.144620 0.354020 In [41]: s = Series(range(5),[-2,-1,1,2,3]) In [42]: s Out[42]: -2 0 -1 1 1 2 2 3 3 4 dtype: int64
Previous Behavior
In [4]: df.loc['2013-01-02':'2013-01-10'] KeyError: 'stop bound [2013-01-10] is not in the [index]' In [6]: s.loc[-10:3] KeyError: 'start bound [-10] is not the [index]'
New Behavior
In [43]: df.loc['2013-01-02':'2013-01-10'] Out[43]: A B C D 2013-01-02 -0.566446 0.036142 -2.074978 0.247792 2013-01-03 -0.897157 -0.136795 0.018289 0.755414 2013-01-04 0.215269 0.841009 -1.445810 -1.401973 2013-01-05 -0.100918 -0.548242 -0.144620 0.354020 In [44]: s.loc[-10:3] �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[44]: -2 0 -1 1 1 2 2 3 3 4 dtype: int64
Allow slicing with float-like values on an integer index for
.ix. Previously this was only enabled for.loc:Previous Behavior
In [8]: s.ix[-1.0:2] TypeError: the slice start value [-1.0] is not a proper indexer for this index type (Int64Index)
New Behavior
In [2]: s.ix[-1.0:2] Out[2]: -1 1 1 2 2 3 dtype: int64
Provide a useful exception for indexing with an invalid type for that index when using
.loc. For example trying to use.locon an index of typeDatetimeIndexorPeriodIndexorTimedeltaIndex, with an integer (or a float).Previous Behavior
In [4]: df.loc[2:3] KeyError: 'start bound [2] is not the [index]'
New Behavior
In [4]: df.loc[2:3] TypeError: Cannot do slice indexing on <class 'pandas.tseries.index.DatetimeIndex'> with <type 'int'> keys
Categorical Changes¶
In prior versions, Categoricals that had an unspecified ordering (meaning no ordered keyword was passed) were defaulted as ordered Categoricals. Going forward, the ordered keyword in the Categorical constructor will default to False. Ordering must now be explicit.
Furthermore, previously you could change the ordered attribute of a Categorical by just setting the attribute, e.g. cat.ordered=True; This is now deprecated and you should use cat.as_ordered() or cat.as_unordered(). These will by default return a new object and not modify the existing object. (GH9347, GH9190)
Previous Behavior
In [3]: s = Series([0,1,2], dtype='category')
In [4]: s
Out[4]:
0 0
1 1
2 2
dtype: category
Categories (3, int64): [0 < 1 < 2]
In [5]: s.cat.ordered
Out[5]: True
In [6]: s.cat.ordered = False
In [7]: s
Out[7]:
0 0
1 1
2 2
dtype: category
Categories (3, int64): [0, 1, 2]
New Behavior
In [45]: s = Series([0,1,2], dtype='category')
In [46]: s
Out[46]:
0 0
1 1
2 2
dtype: category
Categories (3, int64): [0, 1, 2]
In [47]: s.cat.ordered
��������������������������������������������������������������������������������Out[47]: False
In [48]: s = s.cat.as_ordered()
In [49]: s
Out[49]:
0 0
1 1
2 2
dtype: category
Categories (3, int64): [0 < 1 < 2]
In [50]: s.cat.ordered
����������������������������������������������������������������������������������Out[50]: True
# you can set in the constructor of the Categorical
In [51]: s = Series(Categorical([0,1,2],ordered=True))
In [52]: s
Out[52]:
0 0
1 1
2 2
dtype: category
Categories (3, int64): [0 < 1 < 2]
In [53]: s.cat.ordered
����������������������������������������������������������������������������������Out[53]: True
For ease of creation of series of categorical data, we have added the ability to pass keywords when calling .astype(). These are passed directly to the constructor.
In [54]: s = Series(["a","b","c","a"]).astype('category',ordered=True)
In [55]: s
Out[55]:
0 a
1 b
2 c
3 a
dtype: category
Categories (3, object): [a < b < c]
In [56]: s = Series(["a","b","c","a"]).astype('category',categories=list('abcdef'),ordered=False)
In [57]: s
Out[57]:
0 a
1 b
2 c
3 a
dtype: category
Categories (6, object): [a, b, c, d, e, f]
Other API Changes¶
Index.duplicatednow returnsnp.array(dtype=bool)rather thanIndex(dtype=object)containingboolvalues. (GH8875)DataFrame.to_jsonnow returns accurate type serialisation for each column for frames of mixed dtype (GH9037)Previously data was coerced to a common dtype before serialisation, which for example resulted in integers being serialised to floats:
In [2]: pd.DataFrame({'i': [1,2], 'f': [3.0, 4.2]}).to_json() Out[2]: '{"f":{"0":3.0,"1":4.2},"i":{"0":1.0,"1":2.0}}'
Now each column is serialised using its correct dtype:
In [2]: pd.DataFrame({'i': [1,2], 'f': [3.0, 4.2]}).to_json() Out[2]: '{"f":{"0":3.0,"1":4.2},"i":{"0":1,"1":2}}'
DatetimeIndex,PeriodIndexandTimedeltaIndex.summarynow output the same format. (GH9116)TimedeltaIndex.freqstrnow output the same string format asDatetimeIndex. (GH9116)Bar and horizontal bar plots no longer add a dashed line along the info axis. The prior style can be achieved with matplotlib’s
axhlineoraxvlinemethods (GH9088).Seriesaccessors.dt,.catand.strnow raiseAttributeErrorinstead ofTypeErrorif the series does not contain the appropriate type of data (GH9617). This follows Python’s built-in exception hierarchy more closely and ensures that tests likehasattr(s, 'cat')are consistent on both Python 2 and 3.Seriesnow supports bitwise operation for integral types (GH9016). Previously even if the input dtypes were integral, the output dtype was coerced tobool.Previous Behavior
In [2]: pd.Series([0,1,2,3], list('abcd')) | pd.Series([4,4,4,4], list('abcd')) Out[2]: a True b True c True d True dtype: bool
New Behavior. If the input dtypes are integral, the output dtype is also integral and the output values are the result of the bitwise operation.
In [2]: pd.Series([0,1,2,3], list('abcd')) | pd.Series([4,4,4,4], list('abcd')) Out[2]: a 4 b 5 c 6 d 7 dtype: int64
During division involving a
SeriesorDataFrame,0/0and0//0now givenp.naninstead ofnp.inf. (GH9144, GH8445)Previous Behavior
In [2]: p = pd.Series([0, 1]) In [3]: p / 0 Out[3]: 0 inf 1 inf dtype: float64 In [4]: p // 0 Out[4]: 0 inf 1 inf dtype: float64
New Behavior
In [58]: p = pd.Series([0, 1]) In [59]: p / 0 Out[59]: 0 NaN 1 inf dtype: float64 In [60]: p // 0 �������������������������������������������Out[60]: 0 NaN 1 inf dtype: float64
Series.values_countsandSeries.describefor categorical data will now putNaNentries at the end. (GH9443)Series.describefor categorical data will now give counts and frequencies of 0, notNaN, for unused categories (GH9443)Due to a bug fix, looking up a partial string label with
DatetimeIndex.asofnow includes values that match the string, even if they are after the start of the partial string label (GH9258).Old behavior:
In [4]: pd.to_datetime(['2000-01-31', '2000-02-28']).asof('2000-02') Out[4]: Timestamp('2000-01-31 00:00:00')
Fixed behavior:
In [61]: pd.to_datetime(['2000-01-31', '2000-02-28']).asof('2000-02') Out[61]: Timestamp('2000-02-28 00:00:00')
To reproduce the old behavior, simply add more precision to the label (e.g., use
2000-02-01instead of2000-02).
Deprecations¶
- The
rplottrellis plotting interface is deprecated and will be removed in a future version. We refer to external packages like seaborn for similar but more refined functionality (GH3445). The documentation includes some examples how to convert your existing code usingrplotto seaborn: rplot docs. - The
pandas.sandbox.qtpandasinterface is deprecated and will be removed in a future version. We refer users to the external package pandas-qt. (GH9615) - The
pandas.rpyinterface is deprecated and will be removed in a future version. Similar functionaility can be accessed thru the rpy2 project (GH9602) - Adding
DatetimeIndex/PeriodIndexto anotherDatetimeIndex/PeriodIndexis being deprecated as a set-operation. This will be changed to aTypeErrorin a future version..union()should be used for the union set operation. (GH9094) - Subtracting
DatetimeIndex/PeriodIndexfrom anotherDatetimeIndex/PeriodIndexis being deprecated as a set-operation. This will be changed to an actual numeric subtraction yielding aTimeDeltaIndexin a future version..difference()should be used for the differencing set operation. (GH9094)
Removal of prior version deprecations/changes¶
DataFrame.pivot_tableandcrosstab‘srowsandcolskeyword arguments were removed in favor ofindexandcolumns(GH6581)DataFrame.to_excelandDataFrame.to_csvcolskeyword argument was removed in favor ofcolumns(GH6581)- Removed
convert_dummiesin favor ofget_dummies(GH6581) - Removed
value_rangein favor ofdescribe(GH6581)
Performance Improvements¶
- Fixed a performance regression for
.locindexing with an array or list-like (GH9126:). DataFrame.to_json30x performance improvement for mixed dtype frames. (GH9037)- Performance improvements in
MultiIndex.duplicatedby working with labels instead of values (GH9125) - Improved the speed of
nuniqueby callinguniqueinstead ofvalue_counts(GH9129, GH7771) - Performance improvement of up to 10x in
DataFrame.countandDataFrame.dropnaby taking advantage of homogeneous/heterogeneous dtypes appropriately (GH9136) - Performance improvement of up to 20x in
DataFrame.countwhen using aMultiIndexand thelevelkeyword argument (GH9163) - Performance and memory usage improvements in
mergewhen key space exceedsint64bounds (GH9151) - Performance improvements in multi-key
groupby(GH9429) - Performance improvements in
MultiIndex.sortlevel(GH9445) - Performance and memory usage improvements in
DataFrame.duplicated(GH9398) - Cythonized
Period(GH9440) - Decreased memory usage on
to_hdf(GH9648)
Bug Fixes¶
- Changed
.to_htmlto remove leading/trailing spaces in table body (GH4987) - Fixed issue using
read_csvon s3 with Python 3 (GH9452) - Fixed compatibility issue in
DatetimeIndexaffecting architectures wherenumpy.int_defaults tonumpy.int32(GH8943) - Bug in Panel indexing with an object-like (GH9140)
- Bug in the returned
Series.dt.componentsindex was reset to the default index (GH9247) - Bug in
Categorical.__getitem__/__setitem__with listlike input getting incorrect results from indexer coercion (GH9469) - Bug in partial setting with a DatetimeIndex (GH9478)
- Bug in groupby for integer and datetime64 columns when applying an aggregator that caused the value to be changed when the number was sufficiently large (GH9311, GH6620)
- Fixed bug in
to_sqlwhen mapping aTimestampobject column (datetime column with timezone info) to the appropriate sqlalchemy type (GH9085). - Fixed bug in
to_sqldtypeargument not accepting an instantiated SQLAlchemy type (GH9083). - Bug in
.locpartial setting with anp.datetime64(GH9516) - Incorrect dtypes inferred on datetimelike looking
Series& on.xsslices (GH9477) - Items in
Categorical.unique()(ands.unique()ifsis of dtypecategory) now appear in the order in which they are originally found, not in sorted order (GH9331). This is now consistent with the behavior for other dtypes in pandas. - Fixed bug on big endian platforms which produced incorrect results in
StataReader(GH8688). - Bug in
MultiIndex.has_duplicateswhen having many levels causes an indexer overflow (GH9075, GH5873) - Bug in
pivotandunstackwherenanvalues would break index alignment (GH4862, GH7401, GH7403, GH7405, GH7466, GH9497) - Bug in left
joinon multi-index withsort=Trueor null values (GH9210). - Bug in
MultiIndexwhere inserting new keys would fail (GH9250). - Bug in
groupbywhen key space exceedsint64bounds (GH9096). - Bug in
unstackwithTimedeltaIndexorDatetimeIndexand nulls (GH9491). - Bug in
rankwhere comparing floats with tolerance will cause inconsistent behaviour (GH8365). - Fixed character encoding bug in
read_stataandStataReaderwhen loading data from a URL (GH9231). - Bug in adding
offsets.Nanoto other offets raisesTypeError(GH9284) - Bug in
DatetimeIndexiteration, related to (GH8890), fixed in (GH9100) - Bugs in
resamplearound DST transitions. This required fixing offset classes so they behave correctly on DST transitions. (GH5172, GH8744, GH8653, GH9173, GH9468). - Bug in binary operator method (eg
.mul()) alignment with integer levels (GH9463). - Bug in boxplot, scatter and hexbin plot may show an unnecessary warning (GH8877)
- Bug in subplot with
layoutkw may show unnecessary warning (GH9464) - Bug in using grouper functions that need passed thru arguments (e.g. axis), when using wrapped function (e.g.
fillna), (GH9221) DataFramenow properly supports simultaneouscopyanddtypearguments in constructor (GH9099)- Bug in
read_csvwhen using skiprows on a file with CR line endings with the c engine. (GH9079) isnullnow detectsNaTinPeriodIndex(GH9129)- Bug in groupby
.nth()with a multiple column groupby (GH8979) - Bug in
DataFrame.whereandSeries.wherecoerce numerics to string incorrectly (GH9280) - Bug in
DataFrame.whereandSeries.whereraiseValueErrorwhen string list-like is passed. (GH9280) - Accessing
Series.strmethods on with non-string values now raisesTypeErrorinstead of producing incorrect results (GH9184) - Bug in
DatetimeIndex.__contains__when index has duplicates and is not monotonic increasing (GH9512) - Fixed division by zero error for
Series.kurt()when all values are equal (GH9197) - Fixed issue in the
xlsxwriterengine where it added a default ‘General’ format to cells if no other format wass applied. This prevented other row or column formatting being applied. (GH9167) - Fixes issue with
index_col=Falsewhenusecolsis also specified inread_csv. (GH9082) - Bug where
wide_to_longwould modify the input stubnames list (GH9204) - Bug in
to_sqlnot storing float64 values using double precision. (GH9009) SparseSeriesandSparsePanelnow accept zero argument constructors (same as their non-sparse counterparts) (GH9272).- Regression in merging
Categoricalandobjectdtypes (GH9426) - Bug in
read_csvwith buffer overflows with certain malformed input files (GH9205) - Bug in groupby MultiIndex with missing pair (GH9049, GH9344)
- Fixed bug in
Series.groupbywhere grouping onMultiIndexlevels would ignore the sort argument (GH9444) - Fix bug in
DataFrame.Groupbywheresort=Falseis ignored in the case of Categorical columns. (GH8868) - Fixed bug with reading CSV files from Amazon S3 on python 3 raising a TypeError (GH9452)
- Bug in the Google BigQuery reader where the ‘jobComplete’ key may be present but False in the query results (GH8728)
- Bug in
Series.values_countswith excludingNaNfor categorical typeSerieswithdropna=True(GH9443) - Fixed mising numeric_only option for
DataFrame.std/var/sem(GH9201) - Support constructing
PanelorPanel4Dwith scalar data (GH8285) Seriestext representation disconnected from max_rows/max_columns (GH7508).
Seriesnumber formatting inconsistent when truncated (GH8532).Previous Behavior
In [2]: pd.options.display.max_rows = 10 In [3]: s = pd.Series([1,1,1,1,1,1,1,1,1,1,0.9999,1,1]*10) In [4]: s Out[4]: 0 1 1 1 2 1 ... 127 0.9999 128 1.0000 129 1.0000 Length: 130, dtype: float64
New Behavior
0 1.0000 1 1.0000 2 1.0000 3 1.0000 4 1.0000 ... 125 1.0000 126 1.0000 127 0.9999 128 1.0000 129 1.0000 dtype: float64
A Spurious
SettingWithCopyWarning was generated when setting a new item in a frame in some cases (GH8730)The following would previously report a
SettingWithCopyWarning.In [1]: df1 = DataFrame({'x': Series(['a','b','c']), 'y': Series(['d','e','f'])}) In [2]: df2 = df1[['x']] In [3]: df2['y'] = ['g', 'h', 'i']
v0.15.2 (December 12, 2014)¶
This is a minor release from 0.15.1 and includes a large number of bug fixes along with several new features, enhancements, and performance improvements. A small number of API changes were necessary to fix existing bugs. We recommend that all users upgrade to this version.
API changes¶
Indexing in
MultiIndexbeyond lex-sort depth is now supported, though a lexically sorted index will have a better performance. (GH2646)In [1]: df = pd.DataFrame({'jim':[0, 0, 1, 1], ...: 'joe':['x', 'x', 'z', 'y'], ...: 'jolie':np.random.rand(4)}).set_index(['jim', 'joe']) ...: In [2]: df Out[2]: jolie jim joe 0 x 0.123943 x 0.119381 1 z 0.738523 y 0.587304 In [3]: df.index.lexsort_depth ���������������������������������������������������������������������������������������������������������������������Out[3]: 1 # in prior versions this would raise a KeyError # will now show a PerformanceWarning In [4]: df.loc[(1, 'z')] �������������������������������������������������������������������������������������������������������������������������������Out[4]: jolie jim joe 1 z 0.738523 # lexically sorting In [5]: df2 = df.sort_index() In [6]: df2 Out[6]: jolie jim joe 0 x 0.123943 x 0.119381 1 y 0.587304 z 0.738523 In [7]: df2.index.lexsort_depth ���������������������������������������������������������������������������������������������������������������������Out[7]: 2 In [8]: df2.loc[(1,'z')] �������������������������������������������������������������������������������������������������������������������������������Out[8]: jolie jim joe 1 z 0.738523
Bug in unique of Series with
categorydtype, which returned all categories regardless whether they were “used” or not (see GH8559 for the discussion). Previous behaviour was to return all categories:In [3]: cat = pd.Categorical(['a', 'b', 'a'], categories=['a', 'b', 'c']) In [4]: cat Out[4]: [a, b, a] Categories (3, object): [a < b < c] In [5]: cat.unique() Out[5]: array(['a', 'b', 'c'], dtype=object)
Now, only the categories that do effectively occur in the array are returned:
In [9]: cat = pd.Categorical(['a', 'b', 'a'], categories=['a', 'b', 'c']) In [10]: cat.unique() Out[10]: [a, b] Categories (2, object): [a, b]
Series.allandSeries.anynow support thelevelandskipnaparameters.Series.all,Series.any,Index.all, andIndex.anyno longer support theoutandkeepdimsparameters, which existed for compatibility with ndarray. Various index types no longer support theallandanyaggregation functions and will now raiseTypeError. (GH8302).Allow equality comparisons of Series with a categorical dtype and object dtype; previously these would raise
TypeError(GH8938)Bug in
NDFrame: conflicting attribute/column names now behave consistently between getting and setting. Previously, when both a column and attribute namedyexisted,data.ywould return the attribute, whiledata.y = zwould update the column (GH8994)In [11]: data = pd.DataFrame({'x':[1, 2, 3]}) In [12]: data.y = 2 In [13]: data['y'] = [2, 4, 6] In [14]: data Out[14]: x y 0 1 2 1 2 4 2 3 6 # this assignment was inconsistent In [15]: data.y = 5
Old behavior:
In [6]: data.y Out[6]: 2 In [7]: data['y'].values Out[7]: array([5, 5, 5])
New behavior:
In [16]: data.y Out[16]: 5 In [17]: data['y'].values �����������Out[17]: array([2, 4, 6])
Timestamp('now')is now equivalent toTimestamp.now()in that it returns the local time rather than UTC. Also,Timestamp('today')is now equivalent toTimestamp.today()and both havetzas a possible argument. (GH9000)Fix negative step support for label-based slices (GH8753)
Old behavior:
In [1]: s = pd.Series(np.arange(3), ['a', 'b', 'c']) Out[1]: a 0 b 1 c 2 dtype: int64 In [2]: s.loc['c':'a':-1] Out[2]: c 2 dtype: int64
New behavior:
In [18]: s = pd.Series(np.arange(3), ['a', 'b', 'c']) In [19]: s.loc['c':'a':-1] Out[19]: c 2 b 1 a 0 dtype: int64
Enhancements¶
Categorical enhancements:
- Added ability to export Categorical data to Stata (GH8633). See here for limitations of categorical variables exported to Stata data files.
- Added flag
order_categoricalstoStataReaderandread_statato select whether to order imported categorical data (GH8836). See here for more information on importing categorical variables from Stata data files. - Added ability to export Categorical data to to/from HDF5 (GH7621). Queries work the same as if it was an object array. However, the
categorydtyped data is stored in a more efficient manner. See here for an example and caveats w.r.t. prior versions of pandas. - Added support for
searchsorted()on Categorical class (GH8420).
Other enhancements:
Added the ability to specify the SQL type of columns when writing a DataFrame to a database (GH8778). For example, specifying to use the sqlalchemy
Stringtype instead of the defaultTexttype for string columns:from sqlalchemy.types import String data.to_sql('data_dtype', engine, dtype={'Col_1': String})
Series.allandSeries.anynow support thelevelandskipnaparameters (GH8302):In [20]: s = pd.Series([False, True, False], index=[0, 0, 1]) In [21]: s.any(level=0) Out[21]: 0 True 1 False dtype: bool
Panelnow supports theallandanyaggregation functions. (GH8302):In [22]: p = pd.Panel(np.random.rand(2, 5, 4) > 0.1) In [23]: p.all() Out[23]: 0 1 0 True True 1 True True 2 False False 3 True True
Added support for
utcfromtimestamp(),fromtimestamp(), andcombine()on Timestamp class (GH5351).Added Google Analytics (pandas.io.ga) basic documentation (GH8835). See `here<http://pandas.pydata.org/pandas-docs/version/0.15.2/remote_data.html#remote-data-ga>`__.
Timedeltaarithmetic returnsNotImplementedin unknown cases, allowing extensions by custom classes (GH8813).Timedeltanow supports arithemtic withnumpy.ndarrayobjects of the appropriate dtype (numpy 1.8 or newer only) (GH8884).Added
Timedelta.to_timedelta64()method to the public API (GH8884).Added
gbq.generate_bq_schema()function to the gbq module (GH8325).Seriesnow works with map objects the same way as generators (GH8909).Added context manager to
HDFStorefor automatic closing (GH8791).to_datetimegains anexactkeyword to allow for a format to not require an exact match for a provided format string (if itsFalse).exactdefaults toTrue(meaning that exact matching is still the default) (GH8904)Added
axvlinesboolean option to parallel_coordinates plot function, determines whether vertical lines will be printed, default is TrueAdded ability to read table footers to read_html (GH8552)
to_sqlnow infers datatypes of non-NA values for columns that contain NA values and have dtypeobject(GH8778).
Performance¶
Bug Fixes¶
- Bug in concat of Series with
categorydtype which were coercing toobject. (GH8641) - Bug in Timestamp-Timestamp not returning a Timedelta type and datelike-datelike ops with timezones (GH8865)
- Made consistent a timezone mismatch exception (either tz operated with None or incompatible timezone), will now return
TypeErrorrather thanValueError(a couple of edge cases only), (GH8865) - Bug in using a
pd.Grouper(key=...)with no level/axis or level only (GH8795, GH8866) - Report a
TypeErrorwhen invalid/no parameters are passed in a groupby (GH8015) - Bug in packaging pandas with
py2app/cx_Freeze(GH8602, GH8831) - Bug in
groupbysignatures that didn’t include *args or **kwargs (GH8733). io.data.Optionsnow raisesRemoteDataErrorwhen no expiry dates are available from Yahoo and when it receives no data from Yahoo (GH8761), (GH8783).- Unclear error message in csv parsing when passing dtype and names and the parsed data is a different data type (GH8833)
- Bug in slicing a multi-index with an empty list and at least one boolean indexer (GH8781)
io.data.Optionsnow raisesRemoteDataErrorwhen no expiry dates are available from Yahoo (GH8761).Timedeltakwargs may now be numpy ints and floats (GH8757).- Fixed several outstanding bugs for
Timedeltaarithmetic and comparisons (GH8813, GH5963, GH5436). sql_schemanow generates dialect appropriateCREATE TABLEstatements (GH8697)slicestring method now takes step into account (GH8754)- Bug in
BlockManagerwhere setting values with different type would break block integrity (GH8850) - Bug in
DatetimeIndexwhen usingtimeobject as key (GH8667) - Bug in
mergewherehow='left'andsort=Falsewould not preserve left frame order (GH7331) - Bug in
MultiIndex.reindexwhere reindexing at level would not reorder labels (GH4088) - Bug in certain operations with dateutil timezones, manifesting with dateutil 2.3 (GH8639)
- Regression in DatetimeIndex iteration with a Fixed/Local offset timezone (GH8890)
- Bug in
to_datetimewhen parsing a nanoseconds using the%fformat (GH8989) io.data.Optionsnow raisesRemoteDataErrorwhen no expiry dates are available from Yahoo and when it receives no data from Yahoo (GH8761), (GH8783).- Fix: The font size was only set on x axis if vertical or the y axis if horizontal. (GH8765)
- Fixed division by 0 when reading big csv files in python 3 (GH8621)
- Bug in outputing a Multindex with
to_html,index=Falsewhich would add an extra column (GH8452) - Imported categorical variables from Stata files retain the ordinal information in the underlying data (GH8836).
- Defined
.sizeattribute acrossNDFrameobjects to provide compat with numpy >= 1.9.1; buggy withnp.array_split(GH8846) - Skip testing of histogram plots for matplotlib <= 1.2 (GH8648).
- Bug where
get_data_googlereturned object dtypes (GH3995) - Bug in
DataFrame.stack(..., dropna=False)when the DataFrame’scolumnsis aMultiIndexwhoselabelsdo not reference all itslevels. (GH8844) - Bug in that Option context applied on
__enter__(GH8514) - Bug in resample that causes a ValueError when resampling across multiple days and the last offset is not calculated from the start of the range (GH8683)
- Bug where
DataFrame.plot(kind='scatter')fails when checking if an np.array is in the DataFrame (GH8852) - Bug in
pd.infer_freq/DataFrame.inferred_freqthat prevented proper sub-daily frequency inference when the index contained DST days (GH8772). - Bug where index name was still used when plotting a series with
use_index=False(GH8558). - Bugs when trying to stack multiple columns, when some (or all) of the level names are numbers (GH8584).
- Bug in
MultiIndexwhere__contains__returns wrong result if index is not lexically sorted or unique (GH7724) - BUG CSV: fix problem with trailing whitespace in skipped rows, (GH8679), (GH8661), (GH8983)
- Regression in
Timestampdoes not parse ‘Z’ zone designator for UTC (GH8771) - Bug in StataWriter the produces writes strings with 244 characters irrespective of actual size (GH8969)
- Fixed ValueError raised by cummin/cummax when datetime64 Series contains NaT. (GH8965)
- Bug in Datareader returns object dtype if there are missing values (GH8980)
- Bug in plotting if sharex was enabled and index was a timeseries, would show labels on multiple axes (GH3964).
- Bug where passing a unit to the TimedeltaIndex constructor applied the to nano-second conversion twice. (GH9011).
- Bug in plotting of a period-like array (GH9012)
v0.15.1 (November 9, 2014)¶
This is a minor bug-fix release from 0.15.0 and includes a small number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
API changes¶
s.dt.hourand other.dtaccessors will now returnnp.nanfor missing values (rather than previously -1), (GH8689)In [1]: s = Series(date_range('20130101',periods=5,freq='D')) In [2]: s.iloc[2] = np.nan In [3]: s Out[3]: 0 2013-01-01 1 2013-01-02 2 NaT 3 2013-01-04 4 2013-01-05 dtype: datetime64[ns]
previous behavior:
In [6]: s.dt.hour Out[6]: 0 0 1 0 2 -1 3 0 4 0 dtype: int64
current behavior:
In [4]: s.dt.hour Out[4]: 0 0.0 1 0.0 2 NaN 3 0.0 4 0.0 dtype: float64
groupbywithas_index=Falsewill not add erroneous extra columns to result (GH8582):In [5]: np.random.seed(2718281) In [6]: df = pd.DataFrame(np.random.randint(0, 100, (10, 2)), ...: columns=['jim', 'joe']) ...: In [7]: df.head() Out[7]: jim joe 0 61 81 1 96 49 2 55 65 3 72 51 4 77 12 In [8]: ts = pd.Series(5 * np.random.randint(0, 3, 10))
previous behavior:
In [4]: df.groupby(ts, as_index=False).max() Out[4]: NaN jim joe 0 0 72 83 1 5 77 84 2 10 96 65
current behavior:
In [9]: df.groupby(ts, as_index=False).max() Out[9]: jim joe 0 72 83 1 77 84 2 96 65
groupbywill not erroneously exclude columns if the column name conflics with the grouper name (GH8112):In [10]: df = pd.DataFrame({'jim': range(5), 'joe': range(5, 10)}) In [11]: df Out[11]: jim joe 0 0 5 1 1 6 2 2 7 3 3 8 4 4 9 In [12]: gr = df.groupby(df['jim'] < 2)
previous behavior (excludes 1st column from output):
In [4]: gr.apply(sum) Out[4]: joe jim False 24 True 11
current behavior:
In [13]: gr.apply(sum) Out[13]: jim joe jim False 9 24 True 1 11
Support for slicing with monotonic decreasing indexes, even if
startorstopis not found in the index (GH7860):In [14]: s = pd.Series(['a', 'b', 'c', 'd'], [4, 3, 2, 1]) In [15]: s Out[15]: 4 a 3 b 2 c 1 d dtype: object
previous behavior:
In [8]: s.loc[3.5:1.5] KeyError: 3.5
current behavior:
In [16]: s.loc[3.5:1.5] Out[16]: 3 b 2 c dtype: object
io.data.Optionshas been fixed for a change in the format of the Yahoo Options page (GH8612), (GH8741)Note
As a result of a change in Yahoo’s option page layout, when an expiry date is given,
Optionsmethods now return data for a single expiry date. Previously, methods returned all data for the selected month.The
monthandyearparameters have been undeprecated and can be used to get all options data for a given month.If an expiry date that is not valid is given, data for the next expiry after the given date is returned.
Option data frames are now saved on the instance as
callsYYMMDDorputsYYMMDD. Previously they were saved ascallsMMYYandputsMMYY. The next expiry is saved ascallsandputs.New features:
- The expiry parameter can now be a single date or a list-like object containing dates.
- A new property
expiry_dateswas added, which returns all available expiry dates.
Current behavior:
In [17]: from pandas.io.data import Options In [18]: aapl = Options('aapl','yahoo') In [19]: aapl.get_call_data().iloc[0:5,0:1] Out[19]: Last Strike Expiry Type Symbol 80 2014-11-14 call AAPL141114C00080000 29.05 84 2014-11-14 call AAPL141114C00084000 24.80 85 2014-11-14 call AAPL141114C00085000 24.05 86 2014-11-14 call AAPL141114C00086000 22.76 87 2014-11-14 call AAPL141114C00087000 21.74 In [20]: aapl.expiry_dates Out[20]: [datetime.date(2014, 11, 14), datetime.date(2014, 11, 22), datetime.date(2014, 11, 28), datetime.date(2014, 12, 5), datetime.date(2014, 12, 12), datetime.date(2014, 12, 20), datetime.date(2015, 1, 17), datetime.date(2015, 2, 20), datetime.date(2015, 4, 17), datetime.date(2015, 7, 17), datetime.date(2016, 1, 15), datetime.date(2017, 1, 20)] In [21]: aapl.get_near_stock_price(expiry=aapl.expiry_dates[0:3]).iloc[0:5,0:1] Out[21]: Last Strike Expiry Type Symbol 109 2014-11-22 call AAPL141122C00109000 1.48 2014-11-28 call AAPL141128C00109000 1.79 110 2014-11-14 call AAPL141114C00110000 0.55 2014-11-22 call AAPL141122C00110000 1.02 2014-11-28 call AAPL141128C00110000 1.32
- pandas now also registers the
datetime64dtype in matplotlib’s units registry to plot such values as datetimes. This is activated once pandas is imported. In previous versions, plotting an array ofdatetime64values will have resulted in plotted integer values. To keep the previous behaviour, you can dodel matplotlib.units.registry[np.datetime64](GH8614).
Enhancements¶
concatpermits a wider variety of iterables of pandas objects to be passed as the first parameter (GH8645):In [17]: from collections import deque In [18]: df1 = pd.DataFrame([1, 2, 3]) In [19]: df2 = pd.DataFrame([4, 5, 6])
previous behavior:
In [7]: pd.concat(deque((df1, df2))) TypeError: first argument must be a list-like of pandas objects, you passed an object of type "deque"
current behavior:
In [20]: pd.concat(deque((df1, df2))) Out[20]: 0 0 1 1 2 2 3 0 4 1 5 2 6
Represent
MultiIndexlabels with a dtype that utilizes memory based on the level size. In prior versions, the memory usage was a constant 8 bytes per element in each level. In addition, in prior versions, the reported memory usage was incorrect as it didn’t show the usage for the memory occupied by the underling data array. (GH8456)In [21]: dfi = DataFrame(1,index=pd.MultiIndex.from_product([['a'],range(1000)]),columns=['A'])
previous behavior:
# this was underreported in prior versions In [1]: dfi.memory_usage(index=True) Out[1]: Index 8000 # took about 24008 bytes in < 0.15.1 A 8000 dtype: int64
current behavior:
In [22]: dfi.memory_usage(index=True) Out[22]: Index 11040 A 8000 dtype: int64
Added Index properties is_monotonic_increasing and is_monotonic_decreasing (GH8680).
Added option to select columns when importing Stata files (GH7935)
Qualify memory usage in
DataFrame.info()by adding+if it is a lower bound (GH8578)Raise errors in certain aggregation cases where an argument such as
numeric_onlyis not handled (GH8592).Added support for 3-character ISO and non-standard country codes in
io.wb.download()(GH8482)World Bank data requests now will warn/raise based on an
errorsargument, as well as a list of hard-coded country codes and the World Bank’s JSON response. In prior versions, the error messages didn’t look at the World Bank’s JSON response. Problem-inducing input were simply dropped prior to the request. The issue was that many good countries were cropped in the hard-coded approach. All countries will work now, but some bad countries will raise exceptions because some edge cases break the entire response. (GH8482)Added option to
Series.str.split()to return aDataFramerather than aSeries(GH8428)Added option to
df.info(null_counts=None|True|False)to override the default display options and force showing of the null-counts (GH8701)
Bug Fixes¶
- Bug in unpickling of a
CustomBusinessDayobject (GH8591) - Bug in coercing
Categoricalto a records array, e.g.df.to_records()(GH8626) - Bug in
Categoricalnot created properly withSeries.to_frame()(GH8626) - Bug in coercing in astype of a
Categoricalof a passedpd.Categorical(this now raisesTypeErrorcorrectly), (GH8626) - Bug in
cut/qcutwhen usingSeriesandretbins=True(GH8589) - Bug in writing Categorical columns to an SQL database with
to_sql(GH8624). - Bug in comparing
Categoricalof datetime raising when being compared to a scalar datetime (GH8687) - Bug in selecting from a
Categoricalwith.iloc(GH8623) - Bug in groupby-transform with a Categorical (GH8623)
- Bug in duplicated/drop_duplicates with a Categorical (GH8623)
- Bug in
Categoricalreflected comparison operator raising if the first argument was a numpy array scalar (e.g. np.int64) (GH8658) - Bug in Panel indexing with a list-like (GH8710)
- Compat issue is
DataFrame.dtypeswhenoptions.mode.use_inf_as_nullis True (GH8722) - Bug in
read_csv,dialectparameter would not take a string (:issue: 8703) - Bug in slicing a multi-index level with an empty-list (GH8737)
- Bug in numeric index operations of add/sub with Float/Index Index with numpy arrays (GH8608)
- Bug in setitem with empty indexer and unwanted coercion of dtypes (GH8669)
- Bug in ix/loc block splitting on setitem (manifests with integer-like dtypes, e.g. datetime64) (GH8607)
- Bug when doing label based indexing with integers not found in the index for non-unique but monotonic indexes (GH8680).
- Bug when indexing a Float64Index with
np.nanon numpy 1.7 (GH8980). - Fix
shapeattribute forMultiIndex(GH8609) - Bug in
GroupBywhere a name conflict between the grouper and columns would breakgroupbyoperations (GH7115, GH8112) - Fixed a bug where plotting a column
yand specifying a label would mutate the index name of the original DataFrame (GH8494) - Fix regression in plotting of a DatetimeIndex directly with matplotlib (GH8614).
- Bug in
date_rangewhere partially-specified dates would incorporate current date (GH6961) - Bug in Setting by indexer to a scalar value with a mixed-dtype Panel4d was failing (GH8702)
- Bug where
DataReader‘s would fail if one of the symbols passed was invalid. Now returns data for valid symbols and np.nan for invalid (GH8494) - Bug in
get_quote_yahoothat wouldn’t allow non-float return values (GH5229).
v0.15.0 (October 18, 2014)¶
This is a major release from 0.14.1 and includes a small number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
Warning
pandas >= 0.15.0 will no longer support compatibility with NumPy versions < 1.7.0. If you want to use the latest versions of pandas, please upgrade to NumPy >= 1.7.0 (GH7711)
- Highlights include:
- The
Categoricaltype was integrated as a first-class pandas type, see here - New scalar type
Timedelta, and a new index typeTimedeltaIndex, see here - New datetimelike properties accessor
.dtfor Series, see Datetimelike Properties - New DataFrame default display for
df.info()to include memory usage, see Memory Usage read_csvwill now by default ignore blank lines when parsing, see here- API change in using Indexes in set operations, see here
- Enhancements in the handling of timezones, see here
- A lot of improvements to the rolling and expanding moment funtions, see here
- Internal refactoring of the
Indexclass to no longer sub-classndarray, see Internal Refactoring - dropping support for
PyTablesless than version 3.0.0, andnumexprless than version 2.1 (GH7990) - Split indexing documentation into Indexing and Selecting Data and MultiIndex / Advanced Indexing
- Split out string methods documentation into Working with Text Data
- The
- Check the API Changes and deprecations before updating
- Other Enhancements
- Performance Improvements
- Bug Fixes
Warning
In 0.15.0 Index has internally been refactored to no longer sub-class ndarray
but instead subclass PandasObject, similarly to the rest of the pandas objects. This change allows very easy sub-classing and creation of new index types. This should be
a transparent change with only very limited API implications (See the Internal Refactoring)
Warning
The refactorings in Categorical changed the two argument constructor from
“codes/labels and levels” to “values and levels (now called ‘categories’)”. This can lead to subtle bugs. If you use
Categorical directly, please audit your code before updating to this pandas
version and change it to use the from_codes() constructor. See more on Categorical here
New features¶
Categoricals in Series/DataFrame¶
Categorical can now be included in Series and DataFrames and gained new
methods to manipulate. Thanks to Jan Schulz for much of this API/implementation. (GH3943, GH5313, GH5314,
GH7444, GH7839, GH7848, GH7864, GH7914, GH7768, GH8006, GH3678,
GH8075, GH8076, GH8143, GH8453, GH8518).
For full docs, see the categorical introduction and the API documentation.
In [1]: df = DataFrame({"id":[1,2,3,4,5,6], "raw_grade":['a', 'b', 'b', 'a', 'a', 'e']})
In [2]: df["grade"] = df["raw_grade"].astype("category")
In [3]: df["grade"]
Out[3]:
0 a
1 b
2 b
3 a
4 a
5 e
Name: grade, dtype: category
Categories (3, object): [a, b, e]
# Rename the categories
In [4]: df["grade"].cat.categories = ["very good", "good", "very bad"]
# Reorder the categories and simultaneously add the missing categories
In [5]: df["grade"] = df["grade"].cat.set_categories(["very bad", "bad", "medium", "good", "very good"])
In [6]: df["grade"]
Out[6]:
0 very good
1 good
2 good
3 very good
4 very good
5 very bad
Name: grade, dtype: category
Categories (5, object): [very bad, bad, medium, good, very good]
In [7]: df.sort_values("grade")
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[7]:
id raw_grade grade
5 6 e very bad
1 2 b good
2 3 b good
0 1 a very good
3 4 a very good
4 5 a very good
In [8]: df.groupby("grade").size()
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[8]:
grade
very bad 1
bad 0
medium 0
good 2
very good 3
dtype: int64
pandas.core.group_aggandpandas.core.factor_aggwere removed. As an alternative, construct a dataframe and usedf.groupby(<group>).agg(<func>).- Supplying “codes/labels and levels” to the
Categoricalconstructor is not supported anymore. Supplying two arguments to the constructor is now interpreted as “values and levels (now called ‘categories’)”. Please change your code to use thefrom_codes()constructor. - The
Categorical.labelsattribute was renamed toCategorical.codesand is read only. If you want to manipulate codes, please use one of the API methods on Categoricals. - The
Categorical.levelsattribute is renamed toCategorical.categories.
TimedeltaIndex/Scalar¶
We introduce a new scalar type Timedelta, which is a subclass of datetime.timedelta, and behaves in a similar manner,
but allows compatibility with np.timedelta64 types as well as a host of custom representation, parsing, and attributes.
This type is very similar to how Timestamp works for datetimes. It is a nice-API box for the type. See the docs.
(GH3009, GH4533, GH8209, GH8187, GH8190, GH7869, GH7661, GH8345, GH8471)
Warning
Timedelta scalars (and TimedeltaIndex) component fields are not the same as the component fields on a datetime.timedelta object. For example, .seconds on a datetime.timedelta object returns the total number of seconds combined between hours, minutes and seconds. In contrast, the pandas Timedelta breaks out hours, minutes, microseconds and nanoseconds separately.
# Timedelta accessor
In [9]: tds = Timedelta('31 days 5 min 3 sec')
In [10]: tds.minutes
Out[10]: 5L
In [11]: tds.seconds
Out[11]: 3L
# datetime.timedelta accessor
# this is 5 minutes * 60 + 3 seconds
In [12]: tds.to_pytimedelta().seconds
Out[12]: 303
Note: this is no longer true starting from v0.16.0, where full
compatibility with datetime.timedelta is introduced. See the
0.16.0 whatsnew entry
Warning
Prior to 0.15.0 pd.to_timedelta would return a Series for list-like/Series input, and a np.timedelta64 for scalar input.
It will now return a TimedeltaIndex for list-like input, Series for Series input, and Timedelta for scalar input.
The arguments to pd.to_timedelta are now (arg,unit='ns',box=True,coerce=False), previously were (arg,box=True,unit='ns') as these are more logical.
Consruct a scalar
In [9]: Timedelta('1 days 06:05:01.00003')
Out[9]: Timedelta('1 days 06:05:01.000030')
In [10]: Timedelta('15.5us')
��������������������������������������������Out[10]: Timedelta('0 days 00:00:00.000015')
In [11]: Timedelta('1 hour 15.5us')
�����������������������������������������������������������������������������������������Out[11]: Timedelta('0 days 01:00:00.000015')
# negative Timedeltas have this string repr
# to be more consistent with datetime.timedelta conventions
In [12]: Timedelta('-1us')
��������������������������������������������������������������������������������������������������������������������������������������Out[12]: Timedelta('-1 days +23:59:59.999999')
# a NaT
In [13]: Timedelta('nan')
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[13]: NaT
Access fields for a Timedelta
In [14]: td = Timedelta('1 hour 3m 15.5us')
In [15]: td.seconds
Out[15]: 3780
In [16]: td.microseconds
��������������Out[16]: 15
In [17]: td.nanoseconds
��������������������������Out[17]: 500
Construct a TimedeltaIndex
In [18]: TimedeltaIndex(['1 days','1 days, 00:00:05',
....: np.timedelta64(2,'D'),timedelta(days=2,seconds=2)])
....:
Out[18]:
TimedeltaIndex(['1 days 00:00:00', '1 days 00:00:05', '2 days 00:00:00',
'2 days 00:00:02'],
dtype='timedelta64[ns]', freq=None)
Constructing a TimedeltaIndex with a regular range
In [19]: timedelta_range('1 days',periods=5,freq='D')
Out[19]: TimedeltaIndex(['1 days', '2 days', '3 days', '4 days', '5 days'], dtype='timedelta64[ns]', freq='D')
In [20]: timedelta_range(start='1 days',end='2 days',freq='30T')
���������������������������������������������������������������������������������������������������������������Out[20]:
TimedeltaIndex(['1 days 00:00:00', '1 days 00:30:00', '1 days 01:00:00',
'1 days 01:30:00', '1 days 02:00:00', '1 days 02:30:00',
'1 days 03:00:00', '1 days 03:30:00', '1 days 04:00:00',
'1 days 04:30:00', '1 days 05:00:00', '1 days 05:30:00',
'1 days 06:00:00', '1 days 06:30:00', '1 days 07:00:00',
'1 days 07:30:00', '1 days 08:00:00', '1 days 08:30:00',
'1 days 09:00:00', '1 days 09:30:00', '1 days 10:00:00',
'1 days 10:30:00', '1 days 11:00:00', '1 days 11:30:00',
'1 days 12:00:00', '1 days 12:30:00', '1 days 13:00:00',
'1 days 13:30:00', '1 days 14:00:00', '1 days 14:30:00',
'1 days 15:00:00', '1 days 15:30:00', '1 days 16:00:00',
'1 days 16:30:00', '1 days 17:00:00', '1 days 17:30:00',
'1 days 18:00:00', '1 days 18:30:00', '1 days 19:00:00',
'1 days 19:30:00', '1 days 20:00:00', '1 days 20:30:00',
'1 days 21:00:00', '1 days 21:30:00', '1 days 22:00:00',
'1 days 22:30:00', '1 days 23:00:00', '1 days 23:30:00',
'2 days 00:00:00'],
dtype='timedelta64[ns]', freq='30T')
You can now use a TimedeltaIndex as the index of a pandas object
In [21]: s = Series(np.arange(5),
....: index=timedelta_range('1 days',periods=5,freq='s'))
....:
In [22]: s
Out[22]:
1 days 00:00:00 0
1 days 00:00:01 1
1 days 00:00:02 2
1 days 00:00:03 3
1 days 00:00:04 4
Freq: S, dtype: int64
You can select with partial string selections
In [23]: s['1 day 00:00:02']
Out[23]: 2
In [24]: s['1 day':'1 day 00:00:02']
�����������Out[24]:
1 days 00:00:00 0
1 days 00:00:01 1
1 days 00:00:02 2
Freq: S, dtype: int64
Finally, the combination of TimedeltaIndex with DatetimeIndex allow certain combination operations that are NaT preserving:
In [25]: tdi = TimedeltaIndex(['1 days',pd.NaT,'2 days'])
In [26]: tdi.tolist()
Out[26]: [Timedelta('1 days 00:00:00'), NaT, Timedelta('2 days 00:00:00')]
In [27]: dti = date_range('20130101',periods=3)
In [28]: dti.tolist()
Out[28]:
[Timestamp('2013-01-01 00:00:00', freq='D'),
Timestamp('2013-01-02 00:00:00', freq='D'),
Timestamp('2013-01-03 00:00:00', freq='D')]
In [29]: (dti + tdi).tolist()
�������������������������������������������������������������������������������������������������������������������������������������������������Out[29]: [Timestamp('2013-01-02 00:00:00'), NaT, Timestamp('2013-01-05 00:00:00')]
In [30]: (dti - tdi).tolist()
������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[30]: [Timestamp('2012-12-31 00:00:00'), NaT, Timestamp('2013-01-01 00:00:00')]
- iteration of a
Seriese.g.list(Series(...))oftimedelta64[ns]would prior to v0.15.0 returnnp.timedelta64for each element. These will now be wrapped inTimedelta.
Memory Usage¶
Implemented methods to find memory usage of a DataFrame. See the FAQ for more. (GH6852).
A new display option display.memory_usage (see Options and Settings) sets the default behavior of the memory_usage argument in the df.info() method. By default display.memory_usage is True.
In [31]: dtypes = ['int64', 'float64', 'datetime64[ns]', 'timedelta64[ns]',
....: 'complex128', 'object', 'bool']
....:
In [32]: n = 5000
In [33]: data = dict([ (t, np.random.randint(100, size=n).astype(t))
....: for t in dtypes])
....:
In [34]: df = DataFrame(data)
In [35]: df['categorical'] = df['object'].astype('category')
In [36]: df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 5000 entries, 0 to 4999
Data columns (total 8 columns):
bool 5000 non-null bool
complex128 5000 non-null complex128
datetime64[ns] 5000 non-null datetime64[ns]
float64 5000 non-null float64
int64 5000 non-null int64
object 5000 non-null object
timedelta64[ns] 5000 non-null timedelta64[ns]
categorical 5000 non-null category
dtypes: bool(1), category(1), complex128(1), datetime64[ns](1), float64(1), int64(1), object(1), timedelta64[ns](1)
memory usage: 289.1+ KB
Additionally memory_usage() is an available method for a dataframe object which returns the memory usage of each column.
In [37]: df.memory_usage(index=True)
Out[37]:
Index 80
bool 5000
complex128 80000
datetime64[ns] 40000
float64 40000
int64 40000
object 40000
timedelta64[ns] 40000
categorical 10920
dtype: int64
.dt accessor¶
Series has gained an accessor to succinctly return datetime like properties for the values of the Series, if its a datetime/period like Series. (GH7207)
This will return a Series, indexed like the existing Series. See the docs
# datetime
In [38]: s = Series(date_range('20130101 09:10:12',periods=4))
In [39]: s
Out[39]:
0 2013-01-01 09:10:12
1 2013-01-02 09:10:12
2 2013-01-03 09:10:12
3 2013-01-04 09:10:12
dtype: datetime64[ns]
In [40]: s.dt.hour
��������������������������������������������������������������������������������������������������������������������������������Out[40]:
0 9
1 9
2 9
3 9
dtype: int64
In [41]: s.dt.second
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[41]:
0 12
1 12
2 12
3 12
dtype: int64
In [42]: s.dt.day
������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[42]:
0 1
1 2
2 3
3 4
dtype: int64
In [43]: s.dt.freq
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[43]: <Day>
This enables nice expressions like this:
In [44]: s[s.dt.day==2]
Out[44]:
1 2013-01-02 09:10:12
dtype: datetime64[ns]
You can easily produce tz aware transformations:
In [45]: stz = s.dt.tz_localize('US/Eastern')
In [46]: stz
Out[46]:
0 2013-01-01 09:10:12-05:00
1 2013-01-02 09:10:12-05:00
2 2013-01-03 09:10:12-05:00
3 2013-01-04 09:10:12-05:00
dtype: datetime64[ns, US/Eastern]
In [47]: stz.dt.tz
��������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[47]: <DstTzInfo 'US/Eastern' LMT-1 day, 19:04:00 STD>
You can also chain these types of operations:
In [48]: s.dt.tz_localize('UTC').dt.tz_convert('US/Eastern')
Out[48]:
0 2013-01-01 04:10:12-05:00
1 2013-01-02 04:10:12-05:00
2 2013-01-03 04:10:12-05:00
3 2013-01-04 04:10:12-05:00
dtype: datetime64[ns, US/Eastern]
The .dt accessor works for period and timedelta dtypes.
# period
In [49]: s = Series(period_range('20130101',periods=4,freq='D'))
In [50]: s
Out[50]:
0 2013-01-01
1 2013-01-02
2 2013-01-03
3 2013-01-04
dtype: object
In [51]: s.dt.year
������������������������������������������������������������������������������������Out[51]:
0 2013
1 2013
2 2013
3 2013
dtype: int64
In [52]: s.dt.day
���������������������������������������������������������������������������������������������������������������������������������������������������Out[52]:
0 1
1 2
2 3
3 4
dtype: int64
# timedelta
In [53]: s = Series(timedelta_range('1 day 00:00:05',periods=4,freq='s'))
In [54]: s
Out[54]:
0 1 days 00:00:05
1 1 days 00:00:06
2 1 days 00:00:07
3 1 days 00:00:08
dtype: timedelta64[ns]
In [55]: s.dt.days
�����������������������������������������������������������������������������������������������������������������Out[55]:
0 1
1 1
2 1
3 1
dtype: int64
In [56]: s.dt.seconds
��������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[56]:
0 5
1 6
2 7
3 8
dtype: int64
In [57]: s.dt.components
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[57]:
days hours minutes seconds milliseconds microseconds nanoseconds
0 1 0 0 5 0 0 0
1 1 0 0 6 0 0 0
2 1 0 0 7 0 0 0
3 1 0 0 8 0 0 0
Timezone handling improvements¶
tz_localize(None)for tz-awareTimestampandDatetimeIndexnow removes timezone holding local time, previously this resulted inExceptionorTypeError(GH7812)In [58]: ts = Timestamp('2014-08-01 09:00', tz='US/Eastern') In [59]: ts Out[59]: Timestamp('2014-08-01 09:00:00-0400', tz='US/Eastern') In [60]: ts.tz_localize(None) ����������������������������������������������������������������Out[60]: Timestamp('2014-08-01 09:00:00') In [61]: didx = DatetimeIndex(start='2014-08-01 09:00', freq='H', periods=10, tz='US/Eastern') In [62]: didx Out[62]: DatetimeIndex(['2014-08-01 09:00:00-04:00', '2014-08-01 10:00:00-04:00', '2014-08-01 11:00:00-04:00', '2014-08-01 12:00:00-04:00', '2014-08-01 13:00:00-04:00', '2014-08-01 14:00:00-04:00', '2014-08-01 15:00:00-04:00', '2014-08-01 16:00:00-04:00', '2014-08-01 17:00:00-04:00', '2014-08-01 18:00:00-04:00'], dtype='datetime64[ns, US/Eastern]', freq='H') In [63]: didx.tz_localize(None) ����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[63]: DatetimeIndex(['2014-08-01 09:00:00', '2014-08-01 10:00:00', '2014-08-01 11:00:00', '2014-08-01 12:00:00', '2014-08-01 13:00:00', '2014-08-01 14:00:00', '2014-08-01 15:00:00', '2014-08-01 16:00:00', '2014-08-01 17:00:00', '2014-08-01 18:00:00'], dtype='datetime64[ns]', freq='H')
tz_localizenow accepts theambiguouskeyword which allows for passing an array of bools indicating whether the date belongs in DST or not, ‘NaT’ for setting transition times to NaT, ‘infer’ for inferring DST/non-DST, and ‘raise’ (default) for anAmbiguousTimeErrorto be raised. See the docs for more details (GH7943)DataFrame.tz_localizeandDataFrame.tz_convertnow accepts an optionallevelargument for localizing a specific level of a MultiIndex (GH7846)Timestamp.tz_localizeandTimestamp.tz_convertnow raiseTypeErrorin error cases, rather thanException(GH8025)a timeseries/index localized to UTC when inserted into a Series/DataFrame will preserve the UTC timezone (rather than being a naive
datetime64[ns]) asobjectdtype (GH8411)Timestamp.__repr__displaysdateutil.tz.tzoffsetinfo (GH7907)
Rolling/Expanding Moments improvements¶
rolling_min(),rolling_max(),rolling_cov(), androlling_corr()now return objects with allNaNwhenlen(arg) < min_periods <= windowrather than raising. (This makes all rolling functions consistent in this behavior). (GH7766)Prior to 0.15.0
In [64]: s = Series([10, 11, 12, 13])
In [15]: rolling_min(s, window=10, min_periods=5) ValueError: min_periods (5) must be <= window (4)
New behavior
In [4]: pd.rolling_min(s, window=10, min_periods=5) Out[4]: 0 NaN 1 NaN 2 NaN 3 NaN dtype: float64
rolling_max(),rolling_min(),rolling_sum(),rolling_mean(),rolling_median(),rolling_std(),rolling_var(),rolling_skew(),rolling_kurt(),rolling_quantile(),rolling_cov(),rolling_corr(),rolling_corr_pairwise(),rolling_window(), androlling_apply()withcenter=Truepreviously would return a result of the same structure as the inputargwithNaNin the final(window-1)/2entries.Now the final
(window-1)/2entries of the result are calculated as if the inputargwere followed by(window-1)/2NaNvalues (or with shrinking windows, in the case ofrolling_apply()). (GH7925, GH8269)Prior behavior (note final value is
NaN):In [7]: rolling_sum(Series(range(4)), window=3, min_periods=0, center=True) Out[7]: 0 1 1 3 2 6 3 NaN dtype: float64
New behavior (note final value is
5 = sum([2, 3, NaN])):In [7]: rolling_sum(Series(range(4)), window=3, min_periods=0, center=True) Out[7]: 0 1 1 3 2 6 3 5 dtype: float64
rolling_window()now normalizes the weights properly in rolling mean mode (mean=True) so that the calculated weighted means (e.g. ‘triang’, ‘gaussian’) are distributed about the same means as those calculated without weighting (i.e. ‘boxcar’). See the note on normalization for further details. (GH7618)In [65]: s = Series([10.5, 8.8, 11.4, 9.7, 9.3])
Behavior prior to 0.15.0:
In [39]: rolling_window(s, window=3, win_type='triang', center=True) Out[39]: 0 NaN 1 6.583333 2 6.883333 3 6.683333 4 NaN dtype: float64
New behavior
In [10]: pd.rolling_window(s, window=3, win_type='triang', center=True) Out[10]: 0 NaN 1 9.875 2 10.325 3 10.025 4 NaN dtype: float64
Removed
centerargument from allexpanding_functions (see list), as the results produced whencenter=Truedid not make much sense. (GH7925)Added optional
ddofargument toexpanding_cov()androlling_cov(). The default value of1is backwards-compatible. (GH8279)Documented the
ddofargument toexpanding_var(),expanding_std(),rolling_var(), androlling_std(). These functions’ support of addofargument (with a default value of1) was previously undocumented. (GH8064)ewma(),ewmstd(),ewmvol(),ewmvar(),ewmcov(), andewmcorr()now interpretmin_periodsin the same manner that therolling_*()andexpanding_*()functions do: a given result entry will beNaNif the (expanding, in this case) window does not contain at leastmin_periodsvalues. The previous behavior was to set toNaNthemin_periodsentries starting with the first non-NaNvalue. (GH7977)Prior behavior (note values start at index
2, which ismin_periodsafter index0(the index of the first non-empty value)):In [66]: s = Series([1, None, None, None, 2, 3])
In [51]: ewma(s, com=3., min_periods=2) Out[51]: 0 NaN 1 NaN 2 1.000000 3 1.000000 4 1.571429 5 2.189189 dtype: float64
New behavior (note values start at index
4, the location of the 2nd (sincemin_periods=2) non-empty value):In [2]: pd.ewma(s, com=3., min_periods=2) Out[2]: 0 NaN 1 NaN 2 NaN 3 NaN 4 1.759644 5 2.383784 dtype: float64
ewmstd(),ewmvol(),ewmvar(),ewmcov(), andewmcorr()now have an optionaladjustargument, just likeewma()does, affecting how the weights are calculated. The default value ofadjustisTrue, which is backwards-compatible. See Exponentially weighted moment functions for details. (GH7911)ewma(),ewmstd(),ewmvol(),ewmvar(),ewmcov(), andewmcorr()now have an optionalignore_naargument. Whenignore_na=False(the default), missing values are taken into account in the weights calculation. Whenignore_na=True(which reproduces the pre-0.15.0 behavior), missing values are ignored in the weights calculation. (GH7543)In [7]: pd.ewma(Series([None, 1., 8.]), com=2.) Out[7]: 0 NaN 1 1.0 2 5.2 dtype: float64 In [8]: pd.ewma(Series([1., None, 8.]), com=2., ignore_na=True) # pre-0.15.0 behavior Out[8]: 0 1.0 1 1.0 2 5.2 dtype: float64 In [9]: pd.ewma(Series([1., None, 8.]), com=2., ignore_na=False) # new default Out[9]: 0 1.000000 1 1.000000 2 5.846154 dtype: float64
Warning
By default (
ignore_na=False) theewm*()functions’ weights calculation in the presence of missing values is different than in pre-0.15.0 versions. To reproduce the pre-0.15.0 calculation of weights in the presence of missing values one must specify explicitlyignore_na=True.Bug in
expanding_cov(),expanding_corr(),rolling_cov(),rolling_cor(),ewmcov(), andewmcorr()returning results with columns sorted by name and producing an error for non-unique columns; now handles non-unique columns and returns columns in original order (except for the case of two DataFrames withpairwise=False, where behavior is unchanged) (GH7542)Bug in
rolling_count()andexpanding_*()functions unnecessarily producing error message for zero-length data (GH8056)Bug in
rolling_apply()andexpanding_apply()interpretingmin_periods=0asmin_periods=1(GH8080)Bug in
expanding_std()andexpanding_var()for a single value producing a confusing error message (GH7900)Bug in
rolling_std()androlling_var()for a single value producing0rather thanNaN(GH7900)Bug in
ewmstd(),ewmvol(),ewmvar(), andewmcov()calculation of de-biasing factors whenbias=False(the default). Previously an incorrect constant factor was used, based onadjust=True,ignore_na=True, and an infinite number of observations. Now a different factor is used for each entry, based on the actual weights (analogous to the usualN/(N-1)factor). In particular, for a single point a value ofNaNis returned whenbias=False, whereas previously a value of (approximately)0was returned.For example, consider the following pre-0.15.0 results for
ewmvar(..., bias=False), and the corresponding debiasing factors:In [67]: s = Series([1., 2., 0., 4.])
In [89]: ewmvar(s, com=2., bias=False) Out[89]: 0 -2.775558e-16 1 3.000000e-01 2 9.556787e-01 3 3.585799e+00 dtype: float64 In [90]: ewmvar(s, com=2., bias=False) / ewmvar(s, com=2., bias=True) Out[90]: 0 1.25 1 1.25 2 1.25 3 1.25 dtype: float64
Note that entry
0is approximately 0, and the debiasing factors are a constant 1.25. By comparison, the following 0.15.0 results have aNaNfor entry0, and the debiasing factors are decreasing (towards 1.25):In [14]: pd.ewmvar(s, com=2., bias=False) Out[14]: 0 NaN 1 0.500000 2 1.210526 3 4.089069 dtype: float64 In [15]: pd.ewmvar(s, com=2., bias=False) / pd.ewmvar(s, com=2., bias=True) Out[15]: 0 NaN 1 2.083333 2 1.583333 3 1.425439 dtype: float64
See Exponentially weighted moment functions for details. (GH7912)
Improvements in the sql io module¶
Added support for a
chunksizeparameter toto_sqlfunction. This allows DataFrame to be written in chunks and avoid packet-size overflow errors (GH8062).Added support for a
chunksizeparameter toread_sqlfunction. Specifying this argument will return an iterator through chunks of the query result (GH2908).Added support for writing
datetime.dateanddatetime.timeobject columns withto_sql(GH6932).Added support for specifying a
schemato read from/write to withread_sql_tableandto_sql(GH7441, GH7952). For example:df.to_sql('table', engine, schema='other_schema') pd.read_sql_table('table', engine, schema='other_schema')
Added support for writing
NaNvalues withto_sql(GH2754).Added support for writing datetime64 columns with
to_sqlfor all database flavors (GH7103).
Backwards incompatible API changes¶
Breaking changes¶
API changes related to Categorical (see here
for more details):
The
Categoricalconstructor with two arguments changed from “codes/labels and levels” to “values and levels (now called ‘categories’)”. This can lead to subtle bugs. If you useCategoricaldirectly, please audit your code by changing it to use thefrom_codes()constructor.An old function call like (prior to 0.15.0):
pd.Categorical([0,1,0,2,1], levels=['a', 'b', 'c'])
will have to adapted to the following to keep the same behaviour:
In [2]: pd.Categorical.from_codes([0,1,0,2,1], categories=['a', 'b', 'c']) Out[2]: [a, b, a, c, b] Categories (3, object): [a, b, c]
API changes related to the introduction of the Timedelta scalar (see
above for more details):
- Prior to 0.15.0
to_timedelta()would return aSeriesfor list-like/Series input, and anp.timedelta64for scalar input. It will now return aTimedeltaIndexfor list-like input,Seriesfor Series input, andTimedeltafor scalar input.
For API changes related to the rolling and expanding functions, see detailed overview above.
Other notable API changes:
Consistency when indexing with
.locand a list-like indexer when no values are found.In [68]: df = DataFrame([['a'],['b']],index=[1,2]) In [69]: df Out[69]: 0 1 a 2 b
In prior versions there was a difference in these two constructs:
df.loc[[3]]would return a frame reindexed by 3 (with allnp.nanvalues)df.loc[[3],:]would raiseKeyError.
Both will now raise a
KeyError. The rule is that at least 1 indexer must be found when using a list-like and.loc(GH7999)Furthermore in prior versions these were also different:
df.loc[[1,3]]would return a frame reindexed by [1,3]df.loc[[1,3],:]would raiseKeyError.
Both will now return a frame reindex by [1,3]. E.g.
In [70]: df.loc[[1,3]] Out[70]: 0 1 a 3 NaN In [71]: df.loc[[1,3],:] �������������������������������Out[71]: 0 1 a 3 NaN
This can also be seen in multi-axis indexing with a
Panel.In [72]: p = Panel(np.arange(2*3*4).reshape(2,3,4), ....: items=['ItemA','ItemB'], ....: major_axis=[1,2,3], ....: minor_axis=['A','B','C','D']) ....: In [73]: p Out[73]: <class 'pandas.core.panel.Panel'> Dimensions: 2 (items) x 3 (major_axis) x 4 (minor_axis) Items axis: ItemA to ItemB Major_axis axis: 1 to 3 Minor_axis axis: A to D
The following would raise
KeyErrorprior to 0.15.0:In [74]: p.loc[['ItemA','ItemD'],:,'D'] Out[74]: ItemA ItemD 1 3 NaN 2 7 NaN 3 11 NaN
Furthermore,
.locwill raise If no values are found in a multi-index with a list-like indexer:In [75]: s = Series(np.arange(3,dtype='int64'), ....: index=MultiIndex.from_product([['A'],['foo','bar','baz']], ....: names=['one','two']) ....: ).sort_index() ....: In [76]: s Out[76]: one two A bar 1 baz 2 foo 0 dtype: int64 In [77]: try: ....: s.loc[['D']] ....: except KeyError as e: ....: print("KeyError: " + str(e)) ....: ��������������������������������������������������������������������������KeyError: "['D'] not in index"
Assigning values to
Nonenow considers the dtype when choosing an ‘empty’ value (GH7941).Previously, assigning to
Nonein numeric containers changed the dtype to object (or errored, depending on the call). It now usesNaN:In [78]: s = Series([1, 2, 3]) In [79]: s.loc[0] = None In [80]: s Out[80]: 0 NaN 1 2.0 2 3.0 dtype: float64
NaTis now used similarly for datetime containers.For object containers, we now preserve
Nonevalues (previously these were converted toNaNvalues).In [81]: s = Series(["a", "b", "c"]) In [82]: s.loc[0] = None In [83]: s Out[83]: 0 None 1 b 2 c dtype: object
To insert a
NaN, you must explicitly usenp.nan. See the docs.In prior versions, updating a pandas object inplace would not reflect in other python references to this object. (GH8511, GH5104)
In [84]: s = Series([1, 2, 3]) In [85]: s2 = s In [86]: s += 1.5
Behavior prior to v0.15.0
# the original object In [5]: s Out[5]: 0 2.5 1 3.5 2 4.5 dtype: float64 # a reference to the original object In [7]: s2 Out[7]: 0 1 1 2 2 3 dtype: int64
This is now the correct behavior
# the original object In [87]: s Out[87]: 0 2.5 1 3.5 2 4.5 dtype: float64 # a reference to the original object In [88]: s2 ����������������������������������������������������Out[88]: 0 2.5 1 3.5 2 4.5 dtype: float64
Made both the C-based and Python engines for read_csv and read_table ignore empty lines in input as well as whitespace-filled lines, as long as
sepis not whitespace. This is an API change that can be controlled by the keyword parameterskip_blank_lines. See the docs (GH4466)A timeseries/index localized to UTC when inserted into a Series/DataFrame will preserve the UTC timezone and inserted as
objectdtype rather than being converted to a naivedatetime64[ns](GH8411).Bug in passing a
DatetimeIndexwith a timezone that was not being retained in DataFrame construction from a dict (GH7822)In prior versions this would drop the timezone, now it retains the timezone, but gives a column of
objectdtype:In [89]: i = date_range('1/1/2011', periods=3, freq='10s', tz = 'US/Eastern') In [90]: i Out[90]: DatetimeIndex(['2011-01-01 00:00:00-05:00', '2011-01-01 00:00:10-05:00', '2011-01-01 00:00:20-05:00'], dtype='datetime64[ns, US/Eastern]', freq='10S') In [91]: df = DataFrame( {'a' : i } ) In [92]: df Out[92]: a 0 2011-01-01 00:00:00-05:00 1 2011-01-01 00:00:10-05:00 2 2011-01-01 00:00:20-05:00 In [93]: df.dtypes ��������������������������������������������������������������������������������������������������������������������������Out[93]: a datetime64[ns, US/Eastern] dtype: object
Previously this would have yielded a column of
datetime64dtype, but without timezone info.The behaviour of assigning a column to an existing dataframe as df[‘a’] = i remains unchanged (this already returned an
objectcolumn with a timezone).When passing multiple levels to
stack(), it will now raise aValueErrorwhen the levels aren’t all level names or all level numbers (GH7660). See Reshaping by stacking and unstacking.Raise a
ValueErrorindf.to_hdfwith ‘fixed’ format, ifdfhas non-unique columns as the resulting file will be broken (GH7761)SettingWithCopyraise/warnings (according to the optionmode.chained_assignment) will now be issued when setting a value on a sliced mixed-dtype DataFrame using chained-assignment. (GH7845, GH7950)In [1]: df = DataFrame(np.arange(0,9), columns=['count']) In [2]: df['group'] = 'b' In [3]: df.iloc[0:5]['group'] = 'a' /usr/local/bin/ipython:1: SettingWithCopyWarning: A value is trying to be set on a copy of a slice from a DataFrame. Try using .loc[row_indexer,col_indexer] = value instead See the the caveats in the documentation: http://pandas.pydata.org/pandas-docs/stable/indexing.html#indexing-view-versus-copy
merge,DataFrame.merge, andordered_mergenow return the same type as theleftargument (GH7737).Previously an enlargement with a mixed-dtype frame would act unlike
.appendwhich will preserve dtypes (related GH2578, GH8176):In [94]: df = DataFrame([[True, 1],[False, 2]], ....: columns=["female","fitness"]) ....: In [95]: df Out[95]: female fitness 0 True 1 1 False 2 In [96]: df.dtypes �������������������������������������������������������������������Out[96]: female bool fitness int64 dtype: object # dtypes are now preserved In [97]: df.loc[2] = df.loc[1] In [98]: df Out[98]: female fitness 0 True 1 1 False 2 2 False 2 In [99]: df.dtypes ��������������������������������������������������������������������������������������Out[99]: female bool fitness int64 dtype: object
Series.to_csv()now returns a string whenpath=None, matching the behaviour ofDataFrame.to_csv()(GH8215).read_hdfnow raisesIOErrorwhen a file that doesn’t exist is passed in. Previously, a new, empty file was created, and aKeyErrorraised (GH7715).DataFrame.info()now ends its output with a newline character (GH8114)Concatenating no objects will now raise a
ValueErrorrather than a bareException.Merge errors will now be sub-classes of
ValueErrorrather than rawException(GH8501)DataFrame.plotandSeries.plotkeywords are now have consistent orders (GH8037)
Internal Refactoring¶
In 0.15.0 Index has internally been refactored to no longer sub-class ndarray
but instead subclass PandasObject, similarly to the rest of the pandas objects. This
change allows very easy sub-classing and creation of new index types. This should be
a transparent change with only very limited API implications (GH5080, GH7439, GH7796, GH8024, GH8367, GH7997, GH8522):
- you may need to unpickle pandas version < 0.15.0 pickles using
pd.read_picklerather thanpickle.load. See pickle docs - when plotting with a
PeriodIndex, the matplotlib internal axes will now be arrays ofPeriodrather than aPeriodIndex(this is similar to how aDatetimeIndexpasses arrays ofdatetimesnow) - MultiIndexes will now raise similary to other pandas objects w.r.t. truth testing, see here (GH7897).
- When plotting a DatetimeIndex directly with matplotlib’s plot function,
the axis labels will no longer be formatted as dates but as integers (the
internal representation of a
datetime64). UPDATE This is fixed in 0.15.1, see here.
Deprecations¶
- The attributes
Categoricallabelsandlevelsattributes are deprecated and renamed tocodesandcategories. - The
outtypeargument topd.DataFrame.to_dicthas been deprecated in favor oforient. (GH7840) - The
convert_dummiesmethod has been deprecated in favor ofget_dummies(GH8140) - The
infer_dstargument intz_localizewill be deprecated in favor ofambiguousto allow for more flexibility in dealing with DST transitions. Replaceinfer_dst=Truewithambiguous='infer'for the same behavior (GH7943). See the docs for more details. - The top-level
pd.value_rangehas been deprecated and can be replaced by.describe()(GH8481)
The
Indexset operations+and-were deprecated in order to provide these for numeric type operations on certain index types.+can be replaced by.union()or|, and-by.difference(). Further the method nameIndex.diff()is deprecated and can be replaced byIndex.difference()(GH8226)# + Index(['a','b','c']) + Index(['b','c','d']) # should be replaced by Index(['a','b','c']).union(Index(['b','c','d']))
# - Index(['a','b','c']) - Index(['b','c','d']) # should be replaced by Index(['a','b','c']).difference(Index(['b','c','d']))
The
infer_typesargument toread_html()now has no effect and is deprecated (GH7762, GH7032).
Removal of prior version deprecations/changes¶
- Remove
DataFrame.delevelmethod in favor ofDataFrame.reset_index
Enhancements¶
Enhancements in the importing/exporting of Stata files:
- Added support for bool, uint8, uint16 and uint32 datatypes in
to_stata(GH7097, GH7365) - Added conversion option when importing Stata files (GH8527)
DataFrame.to_stataandStataWritercheck string length for compatibility with limitations imposed in dta files where fixed-width strings must contain 244 or fewer characters. Attempting to write Stata dta files with strings longer than 244 characters raises aValueError. (GH7858)read_stataandStataReadercan import missing data information into aDataFrameby setting the argumentconvert_missingtoTrue. When using this options, missing values are returned asStataMissingValueobjects and columns containing missing values haveobjectdata type. (GH8045)
Enhancements in the plotting functions:
- Added
layoutkeyword toDataFrame.plot. You can pass a tuple of(rows, columns), one of which can be-1to automatically infer (GH6667, GH8071). - Allow to pass multiple axes to
DataFrame.plot,histandboxplot(GH5353, GH6970, GH7069) - Added support for
c,colormapandcolorbararguments forDataFrame.plotwithkind='scatter'(GH7780) - Histogram from
DataFrame.plotwithkind='hist'(GH7809), See the docs. - Boxplot from
DataFrame.plotwithkind='box'(GH7998), See the docs.
Other:
read_csvnow has a keyword parameterfloat_precisionwhich specifies which floating-point converter the C engine should use during parsing, see here (GH8002, GH8044)Added
searchsortedmethod toSeriesobjects (GH7447)describe()on mixed-types DataFrames is more flexible. Type-based column filtering is now possible via theinclude/excludearguments. See the docs (GH8164).In [100]: df = DataFrame({'catA': ['foo', 'foo', 'bar'] * 8, .....: 'catB': ['a', 'b', 'c', 'd'] * 6, .....: 'numC': np.arange(24), .....: 'numD': np.arange(24.) + .5}) .....: In [101]: df.describe(include=["object"]) Out[101]: catA catB count 24 24 unique 2 4 top foo b freq 16 6 In [102]: df.describe(include=["number", "object"], exclude=["float"]) ������������������������������������������������������������������������������������������������Out[102]: catA catB numC count 24 24 24.000000 unique 2 4 NaN top foo b NaN freq 16 6 NaN mean NaN NaN 11.500000 std NaN NaN 7.071068 min NaN NaN 0.000000 25% NaN NaN 5.750000 50% NaN NaN 11.500000 75% NaN NaN 17.250000 max NaN NaN 23.000000
Requesting all columns is possible with the shorthand ‘all’
In [103]: df.describe(include='all') Out[103]: catA catB numC numD count 24 24 24.000000 24.000000 unique 2 4 NaN NaN top foo b NaN NaN freq 16 6 NaN NaN mean NaN NaN 11.500000 12.000000 std NaN NaN 7.071068 7.071068 min NaN NaN 0.000000 0.500000 25% NaN NaN 5.750000 6.250000 50% NaN NaN 11.500000 12.000000 75% NaN NaN 17.250000 17.750000 max NaN NaN 23.000000 23.500000
Without those arguments, ‘describe` will behave as before, including only numerical columns or, if none are, only categorical columns. See also the docs
Added
splitas an option to theorientargument inpd.DataFrame.to_dict. (GH7840)The
get_dummiesmethod can now be used on DataFrames. By default only catagorical columns are encoded as 0’s and 1’s, while other columns are left untouched.In [104]: df = DataFrame({'A': ['a', 'b', 'a'], 'B': ['c', 'c', 'b'], .....: 'C': [1, 2, 3]}) .....: In [105]: pd.get_dummies(df) Out[105]: C A_a A_b B_b B_c 0 1 1 0 0 1 1 2 0 1 0 1 2 3 1 0 1 0
PeriodIndexsupportsresolutionas the same asDatetimeIndex(GH7708)pandas.tseries.holidayhas added support for additional holidays and ways to observe holidays (GH7070)pandas.tseries.holiday.Holidaynow supports a list of offsets in Python3 (GH7070)pandas.tseries.holiday.Holidaynow supports a days_of_week parameter (GH7070)GroupBy.nth()now supports selecting multiple nth values (GH7910)In [106]: business_dates = date_range(start='4/1/2014', end='6/30/2014', freq='B') In [107]: df = DataFrame(1, index=business_dates, columns=['a', 'b']) # get the first, 4th, and last date index for each month In [108]: df.groupby((df.index.year, df.index.month)).nth([0, 3, -1]) Out[108]: a b 2014 4 1 1 4 1 1 4 1 1 5 1 1 5 1 1 5 1 1 6 1 1 6 1 1 6 1 1
PeriodandPeriodIndexsupports addition/subtraction withtimedelta-likes (GH7966)If
Periodfreq isD,H,T,S,L,U,N,Timedelta-like can be added if the result can have same freq. Otherwise, only the sameoffsetscan be added.In [109]: idx = pd.period_range('2014-07-01 09:00', periods=5, freq='H') In [110]: idx Out[110]: PeriodIndex(['2014-07-01 09:00', '2014-07-01 10:00', '2014-07-01 11:00', '2014-07-01 12:00', '2014-07-01 13:00'], dtype='period[H]', freq='H') In [111]: idx + pd.offsets.Hour(2) �����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[111]: PeriodIndex(['2014-07-01 11:00', '2014-07-01 12:00', '2014-07-01 13:00', '2014-07-01 14:00', '2014-07-01 15:00'], dtype='period[H]', freq='H') In [112]: idx + Timedelta('120m') ����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[112]: PeriodIndex(['2014-07-01 11:00', '2014-07-01 12:00', '2014-07-01 13:00', '2014-07-01 14:00', '2014-07-01 15:00'], dtype='period[H]', freq='H') In [113]: idx = pd.period_range('2014-07', periods=5, freq='M') In [114]: idx Out[114]: PeriodIndex(['2014-07', '2014-08', '2014-09', '2014-10', '2014-11'], dtype='period[M]', freq='M') In [115]: idx + pd.offsets.MonthEnd(3) ������������������������������������������������������������������������������������������������������������Out[115]: PeriodIndex(['2014-10', '2014-11', '2014-12', '2015-01', '2015-02'], dtype='period[M]', freq='M')
Added experimental compatibility with
openpyxlfor versions >= 2.0. TheDataFrame.to_excelmethodenginekeyword now recognizesopenpyxl1andopenpyxl2which will explicitly require openpyxl v1 and v2 respectively, failing if the requested version is not available. Theopenpyxlengine is a now a meta-engine that automatically uses whichever version of openpyxl is installed. (GH7177)DataFrame.fillnacan now accept aDataFrameas a fill value (GH8377)Passing multiple levels to
stack()will now work when multiple level numbers are passed (GH7660). See Reshaping by stacking and unstacking.set_names(),set_labels(), andset_levels()methods now take an optionallevelkeyword argument to all modification of specific level(s) of a MultiIndex. Additionallyset_names()now accepts a scalar string value when operating on anIndexor on a specific level of aMultiIndex(GH7792)In [116]: idx = MultiIndex.from_product([['a'], range(3), list("pqr")], names=['foo', 'bar', 'baz']) In [117]: idx.set_names('qux', level=0) Out[117]: MultiIndex(levels=[['a'], [0, 1, 2], ['p', 'q', 'r']], labels=[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 1, 1, 1, 2, 2, 2], [0, 1, 2, 0, 1, 2, 0, 1, 2]], names=['qux', 'bar', 'baz']) In [118]: idx.set_names(['qux','baz'], level=[0,1]) ���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[118]: MultiIndex(levels=[['a'], [0, 1, 2], ['p', 'q', 'r']], labels=[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 1, 1, 1, 2, 2, 2], [0, 1, 2, 0, 1, 2, 0, 1, 2]], names=['qux', 'baz', 'baz']) In [119]: idx.set_levels(['a','b','c'], level='bar') ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[119]: MultiIndex(levels=[['a'], ['a', 'b', 'c'], ['p', 'q', 'r']], labels=[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 1, 1, 1, 2, 2, 2], [0, 1, 2, 0, 1, 2, 0, 1, 2]], names=['foo', 'bar', 'baz']) In [120]: idx.set_levels([['a','b','c'],[1,2,3]], level=[1,2]) ���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[120]: MultiIndex(levels=[['a'], ['a', 'b', 'c'], [1, 2, 3]], labels=[[0, 0, 0, 0, 0, 0, 0, 0, 0], [0, 0, 0, 1, 1, 1, 2, 2, 2], [0, 1, 2, 0, 1, 2, 0, 1, 2]], names=['foo', 'bar', 'baz'])
Index.isinnow supports alevelargument to specify which index level to use for membership tests (GH7892, GH7890)In [1]: idx = MultiIndex.from_product([[0, 1], ['a', 'b', 'c']]) In [2]: idx.values Out[2]: array([(0, 'a'), (0, 'b'), (0, 'c'), (1, 'a'), (1, 'b'), (1, 'c')], dtype=object) In [3]: idx.isin(['a', 'c', 'e'], level=1) Out[3]: array([ True, False, True, True, False, True], dtype=bool)
Indexnow supportsduplicatedanddrop_duplicates. (GH4060)In [121]: idx = Index([1, 2, 3, 4, 1, 2]) In [122]: idx Out[122]: Int64Index([1, 2, 3, 4, 1, 2], dtype='int64') In [123]: idx.duplicated() ��������������������������������������������������������Out[123]: array([False, False, False, False, True, True], dtype=bool) In [124]: idx.drop_duplicates() ��������������������������������������������������������������������������������������������������������������������������������Out[124]: Int64Index([1, 2, 3, 4], dtype='int64')
add
copy=Trueargument topd.concatto enable pass thru of complete blocks (GH8252)Added support for numpy 1.8+ data types (
bool_,int_,float_,string_) for conversion to R dataframe (GH8400)
Performance¶
- Performance improvements in
DatetimeIndex.__iter__to allow faster iteration (GH7683) - Performance improvements in
Periodcreation (andPeriodIndexsetitem) (GH5155) - Improvements in Series.transform for significant performance gains (revised) (GH6496)
- Performance improvements in
StataReaderwhen reading large files (GH8040, GH8073) - Performance improvements in
StataWriterwhen writing large files (GH8079) - Performance and memory usage improvements in multi-key
groupby(GH8128) - Performance improvements in groupby
.aggand.applywhere builtins max/min were not mapped to numpy/cythonized versions (GH7722) - Performance improvement in writing to sql (
to_sql) of up to 50% (GH8208). - Performance benchmarking of groupby for large value of ngroups (GH6787)
- Performance improvement in
CustomBusinessDay,CustomBusinessMonth(GH8236) - Performance improvement for
MultiIndex.valuesfor multi-level indexes containing datetimes (GH8543)
Bug Fixes¶
- Bug in pivot_table, when using margins and a dict aggfunc (GH8349)
- Bug in
read_csvwheresqueeze=Truewould return a view (GH8217) - Bug in checking of table name in
read_sqlin certain cases (GH7826). - Bug in
DataFrame.groupbywhereGrouperdoes not recognize level when frequency is specified (GH7885) - Bug in multiindexes dtypes getting mixed up when DataFrame is saved to SQL table (GH8021)
- Bug in
Series0-division with a float and integer operand dtypes (GH7785) - Bug in
Series.astype("unicode")not callingunicodeon the values correctly (GH7758) - Bug in
DataFrame.as_matrix()with mixeddatetime64[ns]andtimedelta64[ns]dtypes (GH7778) - Bug in
HDFStore.select_column()not preserving UTC timezone info when selecting aDatetimeIndex(GH7777) - Bug in
to_datetimewhenformat='%Y%m%d'andcoerce=Trueare specified, where previously an object array was returned (rather than a coerced time-series withNaT), (GH7930) - Bug in
DatetimeIndexandPeriodIndexin-place addition and subtraction cause different result from normal one (GH6527) - Bug in adding and subtracting
PeriodIndexwithPeriodIndexraiseTypeError(GH7741) - Bug in
combine_firstwithPeriodIndexdata raisesTypeError(GH3367) - Bug in multi-index slicing with missing indexers (GH7866)
- Bug in multi-index slicing with various edge cases (GH8132)
- Regression in multi-index indexing with a non-scalar type object (GH7914)
- Bug in
Timestampcomparisons with==andint64dtype (GH8058) - Bug in pickles contains
DateOffsetmay raiseAttributeErrorwhennormalizeattribute is reffered internally (GH7748) - Bug in
Panelwhen usingmajor_xsandcopy=Falseis passed (deprecation warning fails because of missingwarnings) (GH8152). - Bug in pickle deserialization that failed for pre-0.14.1 containers with dup items trying to avoid ambiguity when matching block and manager items, when there’s only one block there’s no ambiguity (GH7794)
- Bug in putting a
PeriodIndexinto aSerieswould convert toint64dtype, rather thanobjectofPeriods(GH7932) - Bug in
HDFStoreiteration when passing a where (GH8014) - Bug in
DataFrameGroupby.transformwhen transforming with a passed non-sorted key (GH8046, GH8430) - Bug in repeated timeseries line and area plot may result in
ValueErroror incorrect kind (GH7733) - Bug in inference in a
MultiIndexwithdatetime.dateinputs (GH7888) - Bug in
getwhere anIndexErrorwould not cause the default value to be returned (GH7725) - Bug in
offsets.apply,rollforwardandrollbackmay reset nanosecond (GH7697) - Bug in
offsets.apply,rollforwardandrollbackmay raiseAttributeErrorifTimestamphasdateutiltzinfo (GH7697) - Bug in sorting a multi-index frame with a
Float64Index(GH8017) - Bug in inconsistent panel setitem with a rhs of a
DataFramefor alignment (GH7763) - Bug in
is_superperiodandis_subperiodcannot handle higher frequencies thanS(GH7760, GH7772, GH7803) - Bug in 32-bit platforms with
Series.shift(GH8129) - Bug in
PeriodIndex.uniquereturns int64np.ndarray(GH7540) - Bug in
groupby.applywith a non-affecting mutation in the function (GH8467) - Bug in
DataFrame.reset_indexwhich hasMultiIndexcontainsPeriodIndexorDatetimeIndexwith tz raisesValueError(GH7746, GH7793) - Bug in
DataFrame.plotwithsubplots=Truemay draw unnecessary minor xticks and yticks (GH7801) - Bug in
StataReaderwhich did not read variable labels in 117 files due to difference between Stata documentation and implementation (GH7816) - Bug in
StataReaderwhere strings were always converted to 244 characters-fixed width irrespective of underlying string size (GH7858) - Bug in
DataFrame.plotandSeries.plotmay ignorerotandfontsizekeywords (GH7844) - Bug in
DatetimeIndex.value_countsdoesn’t preserve tz (GH7735) - Bug in
PeriodIndex.value_countsresults inInt64Index(GH7735) - Bug in
DataFrame.joinwhen doing left join on index and there are multiple matches (GH5391) - Bug in
GroupBy.transform()where int groups with a transform that didn’t preserve the index were incorrectly truncated (GH7972). - Bug in
groupbywhere callable objects without name attributes would take the wrong path, and produce aDataFrameinstead of aSeries(GH7929) - Bug in
groupbyerror message when a DataFrame grouping column is duplicated (GH7511) - Bug in
read_htmlwhere theinfer_typesargument forced coercion of date-likes incorrectly (GH7762, GH7032). - Bug in
Series.str.catwith an index which was filtered as to not include the first item (GH7857) - Bug in
Timestampcannot parsenanosecondfrom string (GH7878) - Bug in
Timestampwith string offset andtzresults incorrect (GH7833) - Bug in
tslib.tz_convertandtslib.tz_convert_singlemay return different results (GH7798) - Bug in
DatetimeIndex.intersectionof non-overlapping timestamps with tz raisesIndexError(GH7880) - Bug in alignment with TimeOps and non-unique indexes (GH8363)
- Bug in
GroupBy.filter()where fast path vs. slow path made the filter return a non scalar value that appeared valid but wasn’t (GH7870). - Bug in
date_range()/DatetimeIndex()when the timezone was inferred from input dates yet incorrect times were returned when crossing DST boundaries (GH7835, GH7901). - Bug in
to_excel()where a negative sign was being prepended to positive infinity and was absent for negative infinity (GH7949) - Bug in area plot draws legend with incorrect
alphawhenstacked=True(GH8027) PeriodandPeriodIndexaddition/subtraction withnp.timedelta64results in incorrect internal representations (GH7740)- Bug in
Holidaywith no offset or observance (GH7987) - Bug in
DataFrame.to_latexformatting when columns or index is aMultiIndex(GH7982). - Bug in
DateOffsetaround Daylight Savings Time produces unexpected results (GH5175). - Bug in
DataFrame.shiftwhere empty columns would throwZeroDivisionErroron numpy 1.7 (GH8019) - Bug in installation where
html_encoding/*.htmlwasn’t installed and therefore some tests were not running correctly (GH7927). - Bug in
read_htmlwherebytesobjects were not tested for in_read(GH7927). - Bug in
DataFrame.stack()when one of the column levels was a datelike (GH8039) - Bug in broadcasting numpy scalars with
DataFrame(GH8116) - Bug in
pivot_tableperformed with namelessindexandcolumnsraisesKeyError(GH8103) - Bug in
DataFrame.plot(kind='scatter')draws points and errorbars with different colors when the color is specified byckeyword (GH8081) - Bug in
Float64Indexwhereiatandatwere not testing and were failing (GH8092). - Bug in
DataFrame.boxplot()where y-limits were not set correctly when producing multiple axes (GH7528, GH5517). - Bug in
read_csvwhere line comments were not handled correctly given a custom line terminator ordelim_whitespace=True(GH8122). - Bug in
read_htmlwhere empty tables caused aStopIteration(GH7575) - Bug in casting when setting a column in a same-dtype block (GH7704)
- Bug in accessing groups from a
GroupBywhen the original grouper was a tuple (GH8121). - Bug in
.atthat would accept integer indexers on a non-integer index and do fallback (GH7814) - Bug with kde plot and NaNs (GH8182)
- Bug in
GroupBy.countwith float32 data type were nan values were not excluded (GH8169). - Bug with stacked barplots and NaNs (GH8175).
- Bug in resample with non evenly divisible offsets (e.g. ‘7s’) (GH8371)
- Bug in interpolation methods with the
limitkeyword when no values needed interpolating (GH7173). - Bug where
col_spacewas ignored inDataFrame.to_string()whenheader=False(GH8230). - Bug with
DatetimeIndex.asofincorrectly matching partial strings and returning the wrong date (GH8245). - Bug in plotting methods modifying the global matplotlib rcParams (GH8242).
- Bug in
DataFrame.__setitem__that caused errors when setting a dataframe column to a sparse array (GH8131) - Bug where
Dataframe.boxplot()failed when entire column was empty (GH8181). - Bug with messed variables in
radvizvisualization (GH8199). - Bug in interpolation methods with the
limitkeyword when no values needed interpolating (GH7173). - Bug where
col_spacewas ignored inDataFrame.to_string()whenheader=False(GH8230). - Bug in
to_clipboardthat would clip long column data (GH8305) - Bug in
DataFrameterminal display: Setting max_column/max_rows to zero did not trigger auto-resizing of dfs to fit terminal width/height (GH7180). - Bug in OLS where running with “cluster” and “nw_lags” parameters did not work correctly, but also did not throw an error (GH5884).
- Bug in
DataFrame.dropnathat interpreted non-existent columns in the subset argument as the ‘last column’ (GH8303) - Bug in
Index.intersectionon non-monotonic non-unique indexes (GH8362). - Bug in masked series assignment where mismatching types would break alignment (GH8387)
- Bug in
NDFrame.equalsgives false negatives with dtype=object (GH8437) - Bug in assignment with indexer where type diversity would break alignment (GH8258)
- Bug in
NDFrame.locindexing when row/column names were lost when target was a list/ndarray (GH6552) - Regression in
NDFrame.locindexing when rows/columns were converted to Float64Index if target was an empty list/ndarray (GH7774) - Bug in
Seriesthat allows it to be indexed by aDataFramewhich has unexpected results. Such indexing is no longer permitted (GH8444) - Bug in item assignment of a
DataFramewith multi-index columns where right-hand-side columns were not aligned (GH7655) - Suppress FutureWarning generated by NumPy when comparing object arrays containing NaN for equality (GH7065)
- Bug in
DataFrame.eval()where the dtype of thenotoperator (~) was not correctly inferred asbool.
v0.14.1 (July 11, 2014)¶
This is a minor release from 0.14.0 and includes a small number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
- Highlights include:
- New methods
select_dtypes()to select columns based on the dtype andsem()to calculate the standard error of the mean. - Support for dateutil timezones (see docs).
- Support for ignoring full line comments in the
read_csv()text parser. - New documentation section on Options and Settings.
- Lots of bug fixes.
- New methods
- Enhancements
- API Changes
- Performance Improvements
- Experimental Changes
- Bug Fixes
API changes¶
Openpyxl now raises a ValueError on construction of the openpyxl writer instead of warning on pandas import (GH7284).
For
StringMethods.extract, when no match is found, the result - only containingNaNvalues - now also hasdtype=objectinstead offloat(GH7242)Periodobjects no longer raise aTypeErrorwhen compared using==with another object that isn’t aPeriod. Instead when comparing aPeriodwith another object using==if the other object isn’t aPeriodFalseis returned. (GH7376)Previously, the behaviour on resetting the time or not in
offsets.apply,rollforwardandrollbackoperations differed between offsets. With the support of thenormalizekeyword for all offsets(see below) with a default value of False (preserve time), the behaviour changed for certain offsets (BusinessMonthBegin, MonthEnd, BusinessMonthEnd, CustomBusinessMonthEnd, BusinessYearBegin, LastWeekOfMonth, FY5253Quarter, LastWeekOfMonth, Easter):In [6]: from pandas.tseries import offsets In [7]: d = pd.Timestamp('2014-01-01 09:00') # old behaviour < 0.14.1 In [8]: d + offsets.MonthEnd() Out[8]: Timestamp('2014-01-31 00:00:00')
Starting from 0.14.1 all offsets preserve time by default. The old behaviour can be obtained with
normalize=True# new behaviour In [1]: d + offsets.MonthEnd() Out[1]: Timestamp('2014-01-31 09:00:00') In [2]: d + offsets.MonthEnd(normalize=True) �����������������������������������������Out[2]: Timestamp('2014-01-31 00:00:00')
Note that for the other offsets the default behaviour did not change.
Add back
#N/A N/Aas a default NA value in text parsing, (regresion from 0.12) (GH5521)Raise a
TypeErroron inplace-setting with a.whereand a nonnp.nanvalue as this is inconsistent with a set-item expression likedf[mask] = None(GH7656)
Enhancements¶
Add
dropnaargument tovalue_countsandnunique(GH5569).Add
select_dtypes()method to allow selection of columns based on dtype (GH7316). See the docs.All
offsetssuppports thenormalizekeyword to specify whetheroffsets.apply,rollforwardandrollbackresets the time (hour, minute, etc) or not (defaultFalse, preserves time) (GH7156):In [3]: import pandas.tseries.offsets as offsets In [4]: day = offsets.Day() In [5]: day.apply(Timestamp('2014-01-01 09:00')) Out[5]: Timestamp('2014-01-02 09:00:00') In [6]: day = offsets.Day(normalize=True) In [7]: day.apply(Timestamp('2014-01-01 09:00')) Out[7]: Timestamp('2014-01-02 00:00:00')
PeriodIndexis represented as the same format asDatetimeIndex(GH7601)StringMethodsnow work on empty Series (GH7242)The file parsers
read_csvandread_tablenow ignore line comments provided by the parameter comment, which accepts only a single character for the C reader. In particular, they allow for comments before file data begins (GH2685)Add
NotImplementedErrorfor simultaneous use ofchunksizeandnrowsfor read_csv() (GH6774).Tests for basic reading of public S3 buckets now exist (GH7281).
read_htmlnow sports anencodingargument that is passed to the underlying parser library. You can use this to read non-ascii encoded web pages (GH7323).read_excelnow supports reading from URLs in the same way thatread_csvdoes. (GH6809)Support for dateutil timezones, which can now be used in the same way as pytz timezones across pandas. (GH4688)
In [8]: rng = date_range('3/6/2012 00:00', periods=10, freq='D', ...: tz='dateutil/Europe/London') ...: In [9]: rng.tz Out[9]: tzfile('/usr/share/zoneinfo/Europe/London')
See the docs.
Implemented
sem(standard error of the mean) operation forSeries,DataFrame,Panel, andGroupby(GH6897)Add
nlargestandnsmallestto theSeriesgroupbywhitelist, which means you can now use these methods on aSeriesGroupByobject (GH7053).All offsets
apply,rollforwardandrollbackcan now handlenp.datetime64, previously results inApplyTypeError(GH7452)PeriodandPeriodIndexcan containNaTin its values (GH7485)Support pickling
Series,DataFrameandPanelobjects with non-unique labels along item axis (index,columnsanditemsrespectively) (GH7370).Improved inference of datetime/timedelta with mixed null objects. Regression from 0.13.1 in interpretation of an object Index with all null elements (GH7431)
Performance¶
- Improvements in dtype inference for numeric operations involving yielding performance gains for dtypes:
int64,timedelta64,datetime64(GH7223) - Improvements in Series.transform for significant performance gains (GH6496)
- Improvements in DataFrame.transform with ufuncs and built-in grouper functions for signifcant performance gains (GH7383)
- Regression in groupby aggregation of datetime64 dtypes (GH7555)
- Improvements in MultiIndex.from_product for large iterables (GH7627)
Experimental¶
pandas.io.data.Optionshas a new method,get_all_datamethod, and now consistently returns a multi-indexedDataFrame(GH5602)io.gbq.read_gbqandio.gbq.to_gbqwere refactored to remove the dependency on the Googlebq.pycommand line client. This submodule now useshttplib2and the Googleapiclientandoauth2clientAPI client libraries which should be more stable and, therefore, reliable thanbq.py. See the docs. (GH6937).
Bug Fixes¶
- Bug in
DataFrame.wherewith a symmetric shaped frame and a passed other of a DataFrame (GH7506) - Bug in Panel indexing with a multi-index axis (GH7516)
- Regression in datetimelike slice indexing with a duplicated index and non-exact end-points (GH7523)
- Bug in setitem with list-of-lists and single vs mixed types (GH7551:)
- Bug in timeops with non-aligned Series (GH7500)
- Bug in timedelta inference when assigning an incomplete Series (GH7592)
- Bug in groupby
.nthwith a Series and integer-like column name (GH7559) - Bug in
Series.getwith a boolean accessor (GH7407) - Bug in
value_countswhereNaTdid not qualify as missing (NaN) (GH7423) - Bug in
to_timedeltathat accepted invalid units and misinterpreted ‘m/h’ (GH7611, GH6423) - Bug in line plot doesn’t set correct
xlimifsecondary_y=True(GH7459) - Bug in grouped
histandscatterplots use oldfigsizedefault (GH7394) - Bug in plotting subplots with
DataFrame.plot,histclears passedaxeven if the number of subplots is one (GH7391). - Bug in plotting subplots with
DataFrame.boxplotwithbykw raisesValueErrorif the number of subplots exceeds 1 (GH7391). - Bug in subplots displays
ticklabelsandlabelsin different rule (GH5897) - Bug in
Panel.applywith a multi-index as an axis (GH7469) - Bug in
DatetimeIndex.insertdoesn’t preservenameandtz(GH7299) - Bug in
DatetimeIndex.asobjectdoesn’t preservename(GH7299) - Bug in multi-index slicing with datetimelike ranges (strings and Timestamps), (GH7429)
- Bug in
Index.minandmaxdoesn’t handlenanandNaTproperly (GH7261) - Bug in
PeriodIndex.min/maxresults inint(GH7609) - Bug in
resamplewherefill_methodwas ignored if you passedhow(GH2073) - Bug in
TimeGrouperdoesn’t exclude column specified bykey(GH7227) - Bug in
DataFrameandSeriesbar and barh plot raisesTypeErrorwhenbottomandleftkeyword is specified (GH7226) - Bug in
DataFrame.histraisesTypeErrorwhen it contains non numeric column (GH7277) - Bug in
Index.deletedoes not preservenameandfreqattributes (GH7302) - Bug in
DataFrame.query()/evalwhere local string variables with the @ sign were being treated as temporaries attempting to be deleted (GH7300). - Bug in
Float64Indexwhich didn’t allow duplicates (GH7149). - Bug in
DataFrame.replace()where truthy values were being replaced (GH7140). - Bug in
StringMethods.extract()where a single match group Series would use the matcher’s name instead of the group name (GH7313). - Bug in
isnull()whenmode.use_inf_as_null == Truewhere isnull wouldn’t testTruewhen it encountered aninf/-inf(GH7315). - Bug in inferred_freq results in None for eastern hemisphere timezones (GH7310)
- Bug in
Easterreturns incorrect date when offset is negative (GH7195) - Bug in broadcasting with
.div, integer dtypes and divide-by-zero (GH7325) - Bug in
CustomBusinessDay.applyraiasesNameErrorwhennp.datetime64object is passed (GH7196) - Bug in
MultiIndex.append,concatandpivot_tabledon’t preserve timezone (GH6606) - Bug in
.locwith a list of indexers on a single-multi index level (that is not nested) (GH7349) - Bug in
Series.mapwhen mapping a dict with tuple keys of different lengths (GH7333) - Bug all
StringMethodsnow work on empty Series (GH7242) - Fix delegation of read_sql to read_sql_query when query does not contain ‘select’ (GH7324).
- Bug where a string column name assignment to a
DataFramewith aFloat64Indexraised aTypeErrorduring a call tonp.isnan(GH7366). - Bug where
NDFrame.replace()didn’t correctly replace objects withPeriodvalues (GH7379). - Bug in
.ixgetitem should always return a Series (GH7150) - Bug in multi-index slicing with incomplete indexers (GH7399)
- Bug in multi-index slicing with a step in a sliced level (GH7400)
- Bug where negative indexers in
DatetimeIndexwere not correctly sliced (GH7408) - Bug where
NaTwasn’t repr’d correctly in aMultiIndex(GH7406, GH7409). - Bug where bool objects were converted to
naninconvert_objects(GH7416). - Bug in
quantileignoring the axis keyword argument (:issue`7306`) - Bug where
nanops._maybe_null_outdoesn’t work with complex numbers (GH7353) - Bug in several
nanopsfunctions whenaxis==0for 1-dimensionalnanarrays (GH7354) - Bug where
nanops.nanmediandoesn’t work whenaxis==None(GH7352) - Bug where
nanops._has_infsdoesn’t work with many dtypes (GH7357) - Bug in
StataReader.datawhere reading a 0-observation dta failed (GH7369) - Bug in
StataReaderwhen reading Stata 13 (117) files containing fixed width strings (GH7360) - Bug in
StataWriterwhere encoding was ignored (GH7286) - Bug in
DatetimeIndexcomparison doesn’t handleNaTproperly (GH7529) - Bug in passing input with
tzinfoto some offsetsapply,rollforwardorrollbackresetstzinfoor raisesValueError(GH7465) - Bug in
DatetimeIndex.to_period,PeriodIndex.asobject,PeriodIndex.to_timestampdoesn’t preservename(GH7485) - Bug in
DatetimeIndex.to_periodandPeriodIndex.to_timestanphandleNaTincorrectly (GH7228) - Bug in
offsets.apply,rollforwardandrollbackmay return normaldatetime(GH7502) - Bug in
resampleraisesValueErrorwhen target containsNaT(GH7227) - Bug in
Timestamp.tz_localizeresetsnanosecondinfo (GH7534) - Bug in
DatetimeIndex.asobjectraisesValueErrorwhen it containsNaT(GH7539) - Bug in
Timestamp.__new__doesn’t preserve nanosecond properly (GH7610) - Bug in
Index.astype(float)where it would return anobjectdtypeIndex(GH7464). - Bug in
DataFrame.reset_indexlosestz(GH3950) - Bug in
DatetimeIndex.freqstrraisesAttributeErrorwhenfreqisNone(GH7606) - Bug in
GroupBy.sizecreated byTimeGrouperraisesAttributeError(GH7453) - Bug in single column bar plot is misaligned (GH7498).
- Bug in area plot with tz-aware time series raises
ValueError(GH7471) - Bug in non-monotonic
Index.unionmay preservenameincorrectly (GH7458) - Bug in
DatetimeIndex.intersectiondoesn’t preserve timezone (GH4690) - Bug in
rolling_varwhere a window larger than the array would raise an error(GH7297) - Bug with last plotted timeseries dictating
xlim(GH2960) - Bug with
secondary_yaxis not being considered for timeseriesxlim(GH3490) - Bug in
Float64Indexassignment with a non scalar indexer (GH7586) - Bug in
pandas.core.strings.str_containsdoes not properly match in a case insensitive fashion whenregex=Falseandcase=False(GH7505) - Bug in
expanding_cov,expanding_corr,rolling_cov, androlling_corrfor two arguments with mismatched index (GH7512) - Bug in
to_sqltaking the boolean column as text column (GH7678) - Bug in grouped hist doesn’t handle rot kw and sharex kw properly (GH7234)
- Bug in
.locperforming fallback integer indexing withobjectdtype indices (GH7496) - Bug (regression) in
PeriodIndexconstructor when passedSeriesobjects (GH7701).
v0.14.0 (May 31 , 2014)¶
This is a major release from 0.13.1 and includes a small number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
- Highlights include:
- Officially support Python 3.4
- SQL interfaces updated to use
sqlalchemy, See Here. - Display interface changes, See Here
- MultiIndexing Using Slicers, See Here.
- Ability to join a singly-indexed DataFrame with a multi-indexed DataFrame, see Here
- More consistency in groupby results and more flexible groupby specifications, See Here
- Holiday calendars are now supported in
CustomBusinessDay, see Here - Several improvements in plotting functions, including: hexbin, area and pie plots, see Here.
- Performance doc section on I/O operations, See Here
- Other Enhancements
- API Changes
- Text Parsing API Changes
- Groupby API Changes
- Performance Improvements
- Prior Deprecations
- Deprecations
- Known Issues
- Bug Fixes
Warning
In 0.14.0 all NDFrame based containers have undergone significant internal refactoring. Before that each block of
homogeneous data had its own labels and extra care was necessary to keep those in sync with the parent container’s labels.
This should not have any visible user/API behavior changes (GH6745)
API changes¶
read_exceluses 0 as the default sheet (GH6573)ilocwill now accept out-of-bounds indexers for slices, e.g. a value that exceeds the length of the object being indexed. These will be excluded. This will make pandas conform more with python/numpy indexing of out-of-bounds values. A single indexer that is out-of-bounds and drops the dimensions of the object will still raiseIndexError(GH6296, GH6299). This could result in an empty axis (e.g. an empty DataFrame being returned)In [1]: dfl = DataFrame(np.random.randn(5,2),columns=list('AB')) In [2]: dfl Out[2]: A B 0 1.583584 -0.438313 1 -0.402537 -0.780572 2 -0.141685 0.542241 3 0.370966 -0.251642 4 0.787484 1.666563 In [3]: dfl.iloc[:,2:3] ���������������������������������������������������������������������������������������������������������������������������������������������Out[3]: Empty DataFrame Columns: [] Index: [0, 1, 2, 3, 4] In [4]: dfl.iloc[:,1:3] ���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[4]: B 0 -0.438313 1 -0.780572 2 0.542241 3 -0.251642 4 1.666563 In [5]: dfl.iloc[4:6] ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[5]: A B 4 0.787484 1.666563
These are out-of-bounds selections
dfl.iloc[[4,5,6]] IndexError: positional indexers are out-of-bounds dfl.iloc[:,4] IndexError: single positional indexer is out-of-bounds
Slicing with negative start, stop & step values handles corner cases better (GH6531):
df.iloc[:-len(df)]is now emptydf.iloc[len(df)::-1]now enumerates all elements in reverse
The
DataFrame.interpolate()keyworddowncastdefault has been changed frominfertoNone. This is to preseve the original dtype unless explicitly requested otherwise (GH6290).When converting a dataframe to HTML it used to return Empty DataFrame. This special case has been removed, instead a header with the column names is returned (GH6062).
SeriesandIndexnow internall share more common operations, e.g.factorize(),nunique(),value_counts()are now supported onIndextypes as well. TheSeries.weekdayproperty from is removed from Series for API consistency. Using aDatetimeIndex/PeriodIndexmethod on a Series will now raise aTypeError. (GH4551, GH4056, GH5519, GH6380, GH7206).Add
is_month_start,is_month_end,is_quarter_start,is_quarter_end,is_year_start,is_year_endaccessors forDateTimeIndex/Timestampwhich return a boolean array of whether the timestamp(s) are at the start/end of the month/quarter/year defined by the frequency of theDateTimeIndex/Timestamp(GH4565, GH6998)Local variable usage has changed in
pandas.eval()/DataFrame.eval()/DataFrame.query()(GH5987). For theDataFramemethods, two things have changed- Column names are now given precedence over locals
- Local variables must be referred to explicitly. This means that even if
you have a local variable that is not a column you must still refer to
it with the
'@'prefix. - You can have an expression like
df.query('@a < a')with no complaints frompandasabout ambiguity of the namea. - The top-level
pandas.eval()function does not allow you use the'@'prefix and provides you with an error message telling you so. NameResolutionErrorwas removed because it isn’t necessary anymore.
Define and document the order of column vs index names in query/eval (GH6676)
concatwill now concatenate mixed Series and DataFrames using the Series name or numbering columns as needed (GH2385). See the docsSlicing and advanced/boolean indexing operations on
Indexclasses as well asIndex.delete()andIndex.drop()methods will no longer change the type of the resulting index (GH6440, GH7040)In [6]: i = pd.Index([1, 2, 3, 'a' , 'b', 'c']) In [7]: i[[0,1,2]] Out[7]: Index([1, 2, 3], dtype='object') In [8]: i.drop(['a', 'b', 'c']) �����������������������������������������Out[8]: Index([1, 2, 3], dtype='object')
Previously, the above operation would return
Int64Index. If you’d like to do this manually, useIndex.astype()In [9]: i[[0,1,2]].astype(np.int_) Out[9]: Int64Index([1, 2, 3], dtype='int64')
set_indexno longer converts MultiIndexes to an Index of tuples. For example, the old behavior returned an Index in this case (GH6459):# Old behavior, casted MultiIndex to an Index In [10]: tuple_ind Out[10]: Index([('a', 'c'), ('a', 'd'), ('b', 'c'), ('b', 'd')], dtype='object') In [11]: df_multi.set_index(tuple_ind) ���������������������������������������������������������������������������������Out[11]: 0 1 (a, c) 0.471435 -1.190976 (a, d) 1.432707 -0.312652 (b, c) -0.720589 0.887163 (b, d) 0.859588 -0.636524 # New behavior In [12]: mi ����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[12]: MultiIndex(levels=[['a', 'b'], ['c', 'd']], labels=[[0, 0, 1, 1], [0, 1, 0, 1]]) In [13]: df_multi.set_index(mi) ����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[13]: 0 1 a c 0.471435 -1.190976 d 1.432707 -0.312652 b c -0.720589 0.887163 d 0.859588 -0.636524
This also applies when passing multiple indices to
set_index:# Old output, 2-level MultiIndex of tuples In [14]: df_multi.set_index([df_multi.index, df_multi.index]) Out[14]: 0 1 (a, c) (a, c) 0.471435 -1.190976 (a, d) (a, d) 1.432707 -0.312652 (b, c) (b, c) -0.720589 0.887163 (b, d) (b, d) 0.859588 -0.636524 # New output, 4-level MultiIndex In [15]: df_multi.set_index([df_multi.index, df_multi.index]) Out[15]: 0 1 a c a c 0.471435 -1.190976 d a d 1.432707 -0.312652 b c b c -0.720589 0.887163 d b d 0.859588 -0.636524
pairwisekeyword was added to the statistical moment functionsrolling_cov,rolling_corr,ewmcov,ewmcorr,expanding_cov,expanding_corrto allow the calculation of moving window covariance and correlation matrices (GH4950). See Computing rolling pairwise covariances and correlations in the docs.In [1]: df = DataFrame(np.random.randn(10,4),columns=list('ABCD')) In [4]: covs = pd.rolling_cov(df[['A','B','C']], df[['B','C','D']], 5, pairwise=True) In [5]: covs[df.index[-1]] Out[5]: B C D A 0.035310 0.326593 -0.505430 B 0.137748 -0.006888 -0.005383 C -0.006888 0.861040 0.020762
Series.iteritems()is now lazy (returns an iterator rather than a list). This was the documented behavior prior to 0.14. (GH6760)Added
nuniqueandvalue_countsfunctions toIndexfor counting unique elements. (GH6734)stackandunstacknow raise aValueErrorwhen thelevelkeyword refers to a non-unique item in theIndex(previously raised aKeyError). (GH6738)drop unused order argument from
Series.sort; args now are in the same order asSeries.order; addna_positionarg to conform toSeries.order(GH6847)default sorting algorithm for
Series.orderis nowquicksort, to conform withSeries.sort(and numpy defaults)add
inplacekeyword toSeries.order/sortto make them inverses (GH6859)DataFrame.sortnow places NaNs at the beginning or end of the sort according to thena_positionparameter. (GH3917)accept
TextFileReaderinconcat, which was affecting a common user idiom (GH6583), this was a regression from 0.13.1Added
factorizefunctions toIndexandSeriesto get indexer and unique values (GH7090)describeon a DataFrame with a mix of Timestamp and string like objects returns a different Index (GH7088). Previously the index was unintentionally sorted.Arithmetic operations with only
booldtypes now give a warning indicating that they are evaluated in Python space for+,-, and*operations and raise for all others (GH7011, GH6762, GH7015, GH7210)x = pd.Series(np.random.rand(10) > 0.5) y = True x + y # warning generated: should do x | y instead x / y # this raises because it doesn't make sense NotImplementedError: operator '/' not implemented for bool dtypes
In
HDFStore,select_as_multiplewill always raise aKeyError, when a key or the selector is not found (GH6177)df['col'] = valueanddf.loc[:,'col'] = valueare now completely equivalent; previously the.locwould not necessarily coerce the dtype of the resultant series (GH6149)dtypesandftypesnow return a series withdtype=objecton empty containers (GH5740)df.to_csvwill now return a string of the CSV data if neither a target path nor a buffer is provided (GH6061)pd.infer_freq()will now raise aTypeErrorif given an invalidSeries/Indextype (GH6407, GH6463)A tuple passed to
DataFame.sort_indexwill be interpreted as the levels of the index, rather than requiring a list of tuple (GH4370)all offset operations now return
Timestamptypes (rather than datetime), Business/Week frequencies were incorrect (GH4069)to_excelnow convertsnp.infinto a string representation, customizable by theinf_repkeyword argument (Excel has no native inf representation) (GH6782)Replace
pandas.compat.scipy.scoreatpercentilewithnumpy.percentile(GH6810).quantileon adatetime[ns]series now returnsTimestampinstead ofnp.datetime64objects (GH6810)change
AssertionErrortoTypeErrorfor invalid types passed toconcat(GH6583)Raise a
TypeErrorwhenDataFrameis passed an iterator as thedataargument (GH5357)
Display Changes¶
The default way of printing large DataFrames has changed. DataFrames exceeding
max_rowsand/ormax_columnsare now displayed in a centrally truncated view, consistent with the printing of apandas.Series(GH5603).In previous versions, a DataFrame was truncated once the dimension constraints were reached and an ellipse (...) signaled that part of the data was cut off.
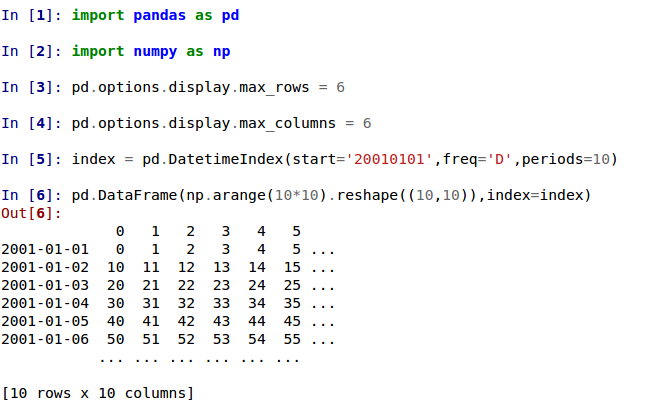
In the current version, large DataFrames are centrally truncated, showing a preview of head and tail in both dimensions.
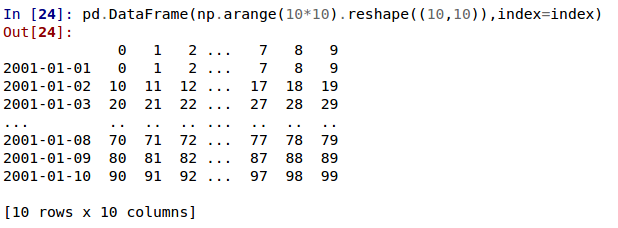
allow option
'truncate'fordisplay.show_dimensionsto only show the dimensions if the frame is truncated (GH6547).The default for
display.show_dimensionswill now betruncate. This is consistent with how Series display length.In [16]: dfd = pd.DataFrame(np.arange(25).reshape(-1,5), index=[0,1,2,3,4], columns=[0,1,2,3,4]) # show dimensions since this is truncated In [17]: with pd.option_context('display.max_rows', 2, 'display.max_columns', 2, ....: 'display.show_dimensions', 'truncate'): ....: print(dfd) ....: 0 ... 4 0 0 ... 4 .. .. ... .. 4 20 ... 24 [5 rows x 5 columns] # will not show dimensions since it is not truncated In [18]: with pd.option_context('display.max_rows', 10, 'display.max_columns', 40, ....: 'display.show_dimensions', 'truncate'): ....: print(dfd) ....: ���������������������������������������������������������������������������������� 0 1 2 3 4 0 0 1 2 3 4 1 5 6 7 8 9 2 10 11 12 13 14 3 15 16 17 18 19 4 20 21 22 23 24
Regression in the display of a MultiIndexed Series with
display.max_rowsis less than the length of the series (GH7101)Fixed a bug in the HTML repr of a truncated Series or DataFrame not showing the class name with the large_repr set to ‘info’ (GH7105)
The verbose keyword in
DataFrame.info(), which controls whether to shorten theinforepresentation, is nowNoneby default. This will follow the global setting indisplay.max_info_columns. The global setting can be overriden withverbose=Trueorverbose=False.Fixed a bug with the info repr not honoring the display.max_info_columns setting (GH6939)
Offset/freq info now in Timestamp __repr__ (GH4553)
Text Parsing API Changes¶
read_csv()/read_table() will now be noiser w.r.t invalid options rather than falling back to the PythonParser.
- Raise
ValueErrorwhensepspecified withdelim_whitespace=Trueinread_csv()/read_table()(GH6607) - Raise
ValueErrorwhenengine='c'specified with unsupported options inread_csv()/read_table()(GH6607) - Raise
ValueErrorwhen fallback to python parser causes options to be ignored (GH6607) - Produce
ParserWarningon fallback to python parser when no options are ignored (GH6607) - Translate
sep='\s+'todelim_whitespace=Trueinread_csv()/read_table()if no other C-unsupported options specified (GH6607)
Groupby API Changes¶
More consistent behaviour for some groupby methods:
groupby
headandtailnow act more likefilterrather than an aggregation:In [19]: df = pd.DataFrame([[1, 2], [1, 4], [5, 6]], columns=['A', 'B']) In [20]: g = df.groupby('A') In [21]: g.head(1) # filters DataFrame Out[21]: A B 0 1 2 2 5 6 In [22]: g.apply(lambda x: x.head(1)) # used to simply fall-through ����������������������������������Out[22]: A B A 1 0 1 2 5 2 5 6
groupby head and tail respect column selection:
In [23]: g[['B']].head(1) Out[23]: B 0 2 2 6
groupby
nthnow reduces by default; filtering can be achieved by passingas_index=False. With an optionaldropnaargument to ignore NaN. See the docs.Reducing
In [24]: df = DataFrame([[1, np.nan], [1, 4], [5, 6]], columns=['A', 'B']) In [25]: g = df.groupby('A') In [26]: g.nth(0) Out[26]: B A 1 NaN 5 6.0 # this is equivalent to g.first() In [27]: g.nth(0, dropna='any') ��������������������������������������Out[27]: B A 1 4.0 5 6.0 # this is equivalent to g.last() In [28]: g.nth(-1, dropna='any') ����������������������������������������������������������������������������Out[28]: B A 1 4.0 5 6.0
Filtering
In [29]: gf = df.groupby('A',as_index=False) In [30]: gf.nth(0) Out[30]: A B 0 1 NaN 2 5 6.0 In [31]: gf.nth(0, dropna='any') ����������������������������������������Out[31]: A B A 1 1 4.0 5 5 6.0
groupby will now not return the grouped column for non-cython functions (GH5610, GH5614, GH6732), as its already the index
In [32]: df = DataFrame([[1, np.nan], [1, 4], [5, 6], [5, 8]], columns=['A', 'B']) In [33]: g = df.groupby('A') In [34]: g.count() Out[34]: B A 1 1 5 2 In [35]: g.describe() ������������������������������Out[35]: B count mean std min 25% 50% 75% max A 1 1.0 4.0 NaN 4.0 4.0 4.0 4.0 4.0 5 2.0 7.0 1.414214 6.0 6.5 7.0 7.5 8.0
passing
as_indexwill leave the grouped column in-place (this is not change in 0.14.0)In [36]: df = DataFrame([[1, np.nan], [1, 4], [5, 6], [5, 8]], columns=['A', 'B']) In [37]: g = df.groupby('A',as_index=False) In [38]: g.count() Out[38]: A B 0 1 1 1 5 2 In [39]: g.describe() ����������������������������������Out[39]: A B \ count mean std min 25% 50% 75% max count mean std min 25% 0 2.0 1.0 0.0 1.0 1.0 1.0 1.0 1.0 1.0 4.0 NaN 4.0 4.0 1 2.0 5.0 0.0 5.0 5.0 5.0 5.0 5.0 2.0 7.0 1.414214 6.0 6.5 50% 75% max 0 4.0 4.0 4.0 1 7.0 7.5 8.0
Allow specification of a more complex groupby via
pd.Grouper, such as grouping by a Time and a string field simultaneously. See the docs. (GH3794)Better propagation/preservation of Series names when performing groupby operations:
SeriesGroupBy.aggwill ensure that the name attribute of the original series is propagated to the result (GH6265).- If the function provided to
GroupBy.applyreturns a named series, the name of the series will be kept as the name of the column index of the DataFrame returned byGroupBy.apply(GH6124). This facilitatesDataFrame.stackoperations where the name of the column index is used as the name of the inserted column containing the pivoted data.
SQL¶
The SQL reading and writing functions now support more database flavors through SQLAlchemy (GH2717, GH4163, GH5950, GH6292). All databases supported by SQLAlchemy can be used, such as PostgreSQL, MySQL, Oracle, Microsoft SQL server (see documentation of SQLAlchemy on included dialects).
The functionality of providing DBAPI connection objects will only be supported
for sqlite3 in the future. The 'mysql' flavor is deprecated.
The new functions read_sql_query() and read_sql_table()
are introduced. The function read_sql() is kept as a convenience
wrapper around the other two and will delegate to specific function depending on
the provided input (database table name or sql query).
In practice, you have to provide a SQLAlchemy engine to the sql functions.
To connect with SQLAlchemy you use the create_engine() function to create an engine
object from database URI. You only need to create the engine once per database you are
connecting to. For an in-memory sqlite database:
In [40]: from sqlalchemy import create_engine
# Create your connection.
In [41]: engine = create_engine('sqlite:///:memory:')
This engine can then be used to write or read data to/from this database:
In [42]: df = pd.DataFrame({'A': [1,2,3], 'B': ['a', 'b', 'c']})
In [43]: df.to_sql('db_table', engine, index=False)
You can read data from a database by specifying the table name:
In [44]: pd.read_sql_table('db_table', engine)
Out[44]:
A B
0 1 a
1 2 b
2 3 c
or by specifying a sql query:
In [45]: pd.read_sql_query('SELECT * FROM db_table', engine)
Out[45]:
A B
0 1 a
1 2 b
2 3 c
Some other enhancements to the sql functions include:
- support for writing the index. This can be controlled with the
indexkeyword (default is True). - specify the column label to use when writing the index with
index_label. - specify string columns to parse as datetimes withh the
parse_dateskeyword inread_sql_query()andread_sql_table().
Warning
Some of the existing functions or function aliases have been deprecated
and will be removed in future versions. This includes: tquery, uquery,
read_frame, frame_query, write_frame.
Warning
The support for the ‘mysql’ flavor when using DBAPI connection objects has been deprecated. MySQL will be further supported with SQLAlchemy engines (GH6900).
MultiIndexing Using Slicers¶
In 0.14.0 we added a new way to slice multi-indexed objects. You can slice a multi-index by providing multiple indexers.
You can provide any of the selectors as if you are indexing by label, see Selection by Label, including slices, lists of labels, labels, and boolean indexers.
You can use slice(None) to select all the contents of that level. You do not need to specify all the
deeper levels, they will be implied as slice(None).
As usual, both sides of the slicers are included as this is label indexing.
See the docs See also issues (GH6134, GH4036, GH3057, GH2598, GH5641, GH7106)
Warning
You should specify all axes in the .loc specifier, meaning the indexer for the index and
for the columns. Their are some ambiguous cases where the passed indexer could be mis-interpreted
as indexing both axes, rather than into say the MuliIndex for the rows.
You should do this:
df.loc[(slice('A1','A3'),.....),:]
rather than this:
df.loc[(slice('A1','A3'),.....)]
Warning
You will need to make sure that the selection axes are fully lexsorted!
In [46]: def mklbl(prefix,n):
....: return ["%s%s" % (prefix,i) for i in range(n)]
....:
In [47]: index = MultiIndex.from_product([mklbl('A',4),
....: mklbl('B',2),
....: mklbl('C',4),
....: mklbl('D',2)])
....:
In [48]: columns = MultiIndex.from_tuples([('a','foo'),('a','bar'),
....: ('b','foo'),('b','bah')],
....: names=['lvl0', 'lvl1'])
....:
In [49]: df = DataFrame(np.arange(len(index)*len(columns)).reshape((len(index),len(columns))),
....: index=index,
....: columns=columns).sort_index().sort_index(axis=1)
....:
In [50]: df
Out[50]:
lvl0 a b
lvl1 bar foo bah foo
A0 B0 C0 D0 1 0 3 2
D1 5 4 7 6
C1 D0 9 8 11 10
D1 13 12 15 14
C2 D0 17 16 19 18
D1 21 20 23 22
C3 D0 25 24 27 26
... ... ... ... ...
A3 B1 C0 D1 229 228 231 230
C1 D0 233 232 235 234
D1 237 236 239 238
C2 D0 241 240 243 242
D1 245 244 247 246
C3 D0 249 248 251 250
D1 253 252 255 254
[64 rows x 4 columns]
Basic multi-index slicing using slices, lists, and labels.
In [51]: df.loc[(slice('A1','A3'),slice(None), ['C1','C3']),:]
Out[51]:
lvl0 a b
lvl1 bar foo bah foo
A1 B0 C1 D0 73 72 75 74
D1 77 76 79 78
C3 D0 89 88 91 90
D1 93 92 95 94
B1 C1 D0 105 104 107 106
D1 109 108 111 110
C3 D0 121 120 123 122
... ... ... ... ...
A3 B0 C1 D1 205 204 207 206
C3 D0 217 216 219 218
D1 221 220 223 222
B1 C1 D0 233 232 235 234
D1 237 236 239 238
C3 D0 249 248 251 250
D1 253 252 255 254
[24 rows x 4 columns]
You can use a pd.IndexSlice to shortcut the creation of these slices
In [52]: idx = pd.IndexSlice
In [53]: df.loc[idx[:,:,['C1','C3']],idx[:,'foo']]
Out[53]:
lvl0 a b
lvl1 foo foo
A0 B0 C1 D0 8 10
D1 12 14
C3 D0 24 26
D1 28 30
B1 C1 D0 40 42
D1 44 46
C3 D0 56 58
... ... ...
A3 B0 C1 D1 204 206
C3 D0 216 218
D1 220 222
B1 C1 D0 232 234
D1 236 238
C3 D0 248 250
D1 252 254
[32 rows x 2 columns]
It is possible to perform quite complicated selections using this method on multiple axes at the same time.
In [54]: df.loc['A1',(slice(None),'foo')]
Out[54]:
lvl0 a b
lvl1 foo foo
B0 C0 D0 64 66
D1 68 70
C1 D0 72 74
D1 76 78
C2 D0 80 82
D1 84 86
C3 D0 88 90
... ... ...
B1 C0 D1 100 102
C1 D0 104 106
D1 108 110
C2 D0 112 114
D1 116 118
C3 D0 120 122
D1 124 126
[16 rows x 2 columns]
In [55]: df.loc[idx[:,:,['C1','C3']],idx[:,'foo']]
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[55]:
lvl0 a b
lvl1 foo foo
A0 B0 C1 D0 8 10
D1 12 14
C3 D0 24 26
D1 28 30
B1 C1 D0 40 42
D1 44 46
C3 D0 56 58
... ... ...
A3 B0 C1 D1 204 206
C3 D0 216 218
D1 220 222
B1 C1 D0 232 234
D1 236 238
C3 D0 248 250
D1 252 254
[32 rows x 2 columns]
Using a boolean indexer you can provide selection related to the values.
In [56]: mask = df[('a','foo')]>200
In [57]: df.loc[idx[mask,:,['C1','C3']],idx[:,'foo']]
Out[57]:
lvl0 a b
lvl1 foo foo
A3 B0 C1 D1 204 206
C3 D0 216 218
D1 220 222
B1 C1 D0 232 234
D1 236 238
C3 D0 248 250
D1 252 254
You can also specify the axis argument to .loc to interpret the passed
slicers on a single axis.
In [58]: df.loc(axis=0)[:,:,['C1','C3']]
Out[58]:
lvl0 a b
lvl1 bar foo bah foo
A0 B0 C1 D0 9 8 11 10
D1 13 12 15 14
C3 D0 25 24 27 26
D1 29 28 31 30
B1 C1 D0 41 40 43 42
D1 45 44 47 46
C3 D0 57 56 59 58
... ... ... ... ...
A3 B0 C1 D1 205 204 207 206
C3 D0 217 216 219 218
D1 221 220 223 222
B1 C1 D0 233 232 235 234
D1 237 236 239 238
C3 D0 249 248 251 250
D1 253 252 255 254
[32 rows x 4 columns]
Furthermore you can set the values using these methods
In [59]: df2 = df.copy()
In [60]: df2.loc(axis=0)[:,:,['C1','C3']] = -10
In [61]: df2
Out[61]:
lvl0 a b
lvl1 bar foo bah foo
A0 B0 C0 D0 1 0 3 2
D1 5 4 7 6
C1 D0 -10 -10 -10 -10
D1 -10 -10 -10 -10
C2 D0 17 16 19 18
D1 21 20 23 22
C3 D0 -10 -10 -10 -10
... ... ... ... ...
A3 B1 C0 D1 229 228 231 230
C1 D0 -10 -10 -10 -10
D1 -10 -10 -10 -10
C2 D0 241 240 243 242
D1 245 244 247 246
C3 D0 -10 -10 -10 -10
D1 -10 -10 -10 -10
[64 rows x 4 columns]
You can use a right-hand-side of an alignable object as well.
In [62]: df2 = df.copy()
In [63]: df2.loc[idx[:,:,['C1','C3']],:] = df2*1000
In [64]: df2
Out[64]:
lvl0 a b
lvl1 bar foo bah foo
A0 B0 C0 D0 1 0 3 2
D1 5 4 7 6
C1 D0 9000 8000 11000 10000
D1 13000 12000 15000 14000
C2 D0 17 16 19 18
D1 21 20 23 22
C3 D0 25000 24000 27000 26000
... ... ... ... ...
A3 B1 C0 D1 229 228 231 230
C1 D0 233000 232000 235000 234000
D1 237000 236000 239000 238000
C2 D0 241 240 243 242
D1 245 244 247 246
C3 D0 249000 248000 251000 250000
D1 253000 252000 255000 254000
[64 rows x 4 columns]
Plotting¶
Hexagonal bin plots from
DataFrame.plotwithkind='hexbin'(GH5478), See the docs.DataFrame.plotandSeries.plotnow supports area plot with specifyingkind='area'(GH6656), See the docsPie plots from
Series.plotandDataFrame.plotwithkind='pie'(GH6976), See the docs.Plotting with Error Bars is now supported in the
.plotmethod ofDataFrameandSeriesobjects (GH3796, GH6834), See the docs.DataFrame.plotandSeries.plotnow support atablekeyword for plottingmatplotlib.Table, See the docs. Thetablekeyword can receive the following values.False: Do nothing (default).True: Draw a table using theDataFrameorSeriescalledplotmethod. Data will be transposed to meet matplotlib’s default layout.DataFrameorSeries: Draw matplotlib.table using the passed data. The data will be drawn as displayed in print method (not transposed automatically). Also, helper functionpandas.tools.plotting.tableis added to create a table fromDataFrameandSeries, and add it to anmatplotlib.Axes.
plot(legend='reverse')will now reverse the order of legend labels for most plot kinds. (GH6014)Line plot and area plot can be stacked by
stacked=True(GH6656)Following keywords are now acceptable for
DataFrame.plot()withkind='bar'andkind='barh':- width: Specify the bar width. In previous versions, static value 0.5 was passed to matplotlib and it cannot be overwritten. (GH6604)
- align: Specify the bar alignment. Default is center (different from matplotlib). In previous versions, pandas passes align=’edge’ to matplotlib and adjust the location to center by itself, and it results align keyword is not applied as expected. (GH4525)
- position: Specify relative alignments for bar plot layout. From 0 (left/bottom-end) to 1(right/top-end). Default is 0.5 (center). (GH6604)
Because of the default align value changes, coordinates of bar plots are now located on integer values (0.0, 1.0, 2.0 ...). This is intended to make bar plot be located on the same coodinates as line plot. However, bar plot may differs unexpectedly when you manually adjust the bar location or drawing area, such as using set_xlim, set_ylim, etc. In this cases, please modify your script to meet with new coordinates.
The
parallel_coordinates()function now takes argumentcolorinstead ofcolors. AFutureWarningis raised to alert that the oldcolorsargument will not be supported in a future release. (GH6956)The
parallel_coordinates()andandrews_curves()functions now take positional argumentframeinstead ofdata. AFutureWarningis raised if the olddataargument is used by name. (GH6956)DataFrame.boxplot()now supportslayoutkeyword (GH6769)DataFrame.boxplot()has a new keyword argument, return_type. It accepts'dict','axes', or'both', in which case a namedtuple with the matplotlib axes and a dict of matplotlib Lines is returned.
Prior Version Deprecations/Changes¶
There are prior version deprecations that are taking effect as of 0.14.0.
- Remove
DateRangein favor ofDatetimeIndex(GH6816) - Remove
columnkeyword fromDataFrame.sort(GH4370) - Remove
precisionkeyword fromset_eng_float_format()(GH395) - Remove
force_unicodekeyword fromDataFrame.to_string(),DataFrame.to_latex(), andDataFrame.to_html(); these function encode in unicode by default (GH2224, GH2225) - Remove
nanRepkeyword fromDataFrame.to_csv()andDataFrame.to_string()(GH275) - Remove
uniquekeyword fromHDFStore.select_column()(GH3256) - Remove
inferTimeRulekeyword fromTimestamp.offset()(GH391) - Remove
namekeyword fromget_data_yahoo()andget_data_google()( commit b921d1a ) - Remove
offsetkeyword fromDatetimeIndexconstructor ( commit 3136390 ) - Remove
time_rulefrom several rolling-moment statistical functions, such asrolling_sum()(GH1042) - Removed neg
-boolean operations on numpy arrays in favor of inv~, as this is going to be deprecated in numpy 1.9 (GH6960)
Deprecations¶
The
pivot_table()/DataFrame.pivot_table()andcrosstab()functions now take argumentsindexandcolumnsinstead ofrowsandcols. AFutureWarningis raised to alert that the oldrowsandcolsarguments will not be supported in a future release (GH5505)The
DataFrame.drop_duplicates()andDataFrame.duplicated()methods now take argumentsubsetinstead ofcolsto better align withDataFrame.dropna(). AFutureWarningis raised to alert that the oldcolsarguments will not be supported in a future release (GH6680)The
DataFrame.to_csv()andDataFrame.to_excel()functions now takes argumentcolumnsinstead ofcols. AFutureWarningis raised to alert that the oldcolsarguments will not be supported in a future release (GH6645)Indexers will warn
FutureWarningwhen used with a scalar indexer and a non-floating point Index (GH4892, GH6960)# non-floating point indexes can only be indexed by integers / labels In [1]: Series(1,np.arange(5))[3.0] pandas/core/index.py:469: FutureWarning: scalar indexers for index type Int64Index should be integers and not floating point Out[1]: 1 In [2]: Series(1,np.arange(5)).iloc[3.0] pandas/core/index.py:469: FutureWarning: scalar indexers for index type Int64Index should be integers and not floating point Out[2]: 1 In [3]: Series(1,np.arange(5)).iloc[3.0:4] pandas/core/index.py:527: FutureWarning: slice indexers when using iloc should be integers and not floating point Out[3]: 3 1 dtype: int64 # these are Float64Indexes, so integer or floating point is acceptable In [4]: Series(1,np.arange(5.))[3] Out[4]: 1 In [5]: Series(1,np.arange(5.))[3.0] Out[6]: 1
Numpy 1.9 compat w.r.t. deprecation warnings (GH6960)
Panel.shift()now has a function signature that matchesDataFrame.shift(). The old positional argumentlagshas been changed to a keyword argumentperiodswith a default value of 1. AFutureWarningis raised if the old argumentlagsis used by name. (GH6910)The
orderkeyword argument offactorize()will be removed. (GH6926).Remove the
copykeyword fromDataFrame.xs(),Panel.major_xs(),Panel.minor_xs(). A view will be returned if possible, otherwise a copy will be made. Previously the user could think thatcopy=Falsewould ALWAYS return a view. (GH6894)The
parallel_coordinates()function now takes argumentcolorinstead ofcolors. AFutureWarningis raised to alert that the oldcolorsargument will not be supported in a future release. (GH6956)The
parallel_coordinates()andandrews_curves()functions now take positional argumentframeinstead ofdata. AFutureWarningis raised if the olddataargument is used by name. (GH6956)The support for the ‘mysql’ flavor when using DBAPI connection objects has been deprecated. MySQL will be further supported with SQLAlchemy engines (GH6900).
The following
io.sqlfunctions have been deprecated:tquery,uquery,read_frame,frame_query,write_frame.The percentile_width keyword argument in
describe()has been deprecated. Use the percentiles keyword instead, which takes a list of percentiles to display. The default output is unchanged.The default return type of
boxplot()will change from a dict to a matpltolib Axes in a future release. You can use the future behavior now by passingreturn_type='axes'to boxplot.
Enhancements¶
DataFrame and Series will create a MultiIndex object if passed a tuples dict, See the docs (GH3323)
In [65]: Series({('a', 'b'): 1, ('a', 'a'): 0, ....: ('a', 'c'): 2, ('b', 'a'): 3, ('b', 'b'): 4}) ....: Out[65]: a a 0 b 1 c 2 b a 3 b 4 dtype: int64 In [66]: DataFrame({('a', 'b'): {('A', 'B'): 1, ('A', 'C'): 2}, ....: ('a', 'a'): {('A', 'C'): 3, ('A', 'B'): 4}, ....: ('a', 'c'): {('A', 'B'): 5, ('A', 'C'): 6}, ....: ('b', 'a'): {('A', 'C'): 7, ('A', 'B'): 8}, ....: ('b', 'b'): {('A', 'D'): 9, ('A', 'B'): 10}}) ....: �������������������������������������������������������������������������Out[66]: a b a b c a b A B 4.0 1.0 5.0 8.0 10.0 C 3.0 2.0 6.0 7.0 NaN D NaN NaN NaN NaN 9.0
Added the
sym_diffmethod toIndex(GH5543)DataFrame.to_latexnow takes a longtable keyword, which if True will return a table in a longtable environment. (GH6617)Add option to turn off escaping in
DataFrame.to_latex(GH6472)pd.read_clipboardwill, if the keywordsepis unspecified, try to detect data copied from a spreadsheet and parse accordingly. (GH6223)Joining a singly-indexed DataFrame with a multi-indexed DataFrame (GH3662)
See the docs. Joining multi-index DataFrames on both the left and right is not yet supported ATM.
In [67]: household = DataFrame(dict(household_id = [1,2,3], ....: male = [0,1,0], ....: wealth = [196087.3,316478.7,294750]), ....: columns = ['household_id','male','wealth'] ....: ).set_index('household_id') ....: In [68]: household Out[68]: male wealth household_id 1 0 196087.3 2 1 316478.7 3 0 294750.0 In [69]: portfolio = DataFrame(dict(household_id = [1,2,2,3,3,3,4], ....: asset_id = ["nl0000301109","nl0000289783","gb00b03mlx29", ....: "gb00b03mlx29","lu0197800237","nl0000289965",np.nan], ....: name = ["ABN Amro","Robeco","Royal Dutch Shell","Royal Dutch Shell", ....: "AAB Eastern Europe Equity Fund","Postbank BioTech Fonds",np.nan], ....: share = [1.0,0.4,0.6,0.15,0.6,0.25,1.0]), ....: columns = ['household_id','asset_id','name','share'] ....: ).set_index(['household_id','asset_id']) ....: In [70]: portfolio Out[70]: name share household_id asset_id 1 nl0000301109 ABN Amro 1.00 2 nl0000289783 Robeco 0.40 gb00b03mlx29 Royal Dutch Shell 0.60 3 gb00b03mlx29 Royal Dutch Shell 0.15 lu0197800237 AAB Eastern Europe Equity Fund 0.60 nl0000289965 Postbank BioTech Fonds 0.25 4 NaN NaN 1.00 In [71]: household.join(portfolio, how='inner') �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[71]: male wealth name \ household_id asset_id 1 nl0000301109 0 196087.3 ABN Amro 2 nl0000289783 1 316478.7 Robeco gb00b03mlx29 1 316478.7 Royal Dutch Shell 3 gb00b03mlx29 0 294750.0 Royal Dutch Shell lu0197800237 0 294750.0 AAB Eastern Europe Equity Fund nl0000289965 0 294750.0 Postbank BioTech Fonds share household_id asset_id 1 nl0000301109 1.00 2 nl0000289783 0.40 gb00b03mlx29 0.60 3 gb00b03mlx29 0.15 lu0197800237 0.60 nl0000289965 0.25
quotechar,doublequote, andescapecharcan now be specified when usingDataFrame.to_csv(GH5414, GH4528)Partially sort by only the specified levels of a MultiIndex with the
sort_remainingboolean kwarg. (GH3984)Added
to_julian_datetoTimeStampandDatetimeIndex. The Julian Date is used primarily in astronomy and represents the number of days from noon, January 1, 4713 BC. Because nanoseconds are used to define the time in pandas the actual range of dates that you can use is 1678 AD to 2262 AD. (GH4041)DataFrame.to_statawill now check data for compatibility with Stata data types and will upcast when needed. When it is not possible to losslessly upcast, a warning is issued (GH6327)DataFrame.to_stataandStataWriterwill accept keyword arguments time_stamp and data_label which allow the time stamp and dataset label to be set when creating a file. (GH6545)pandas.io.gbqnow handles reading unicode strings properly. (GH5940)Holidays Calendars are now available and can be used with the
CustomBusinessDayoffset (GH6719)Float64Indexis now backed by afloat64dtype ndarray instead of anobjectdtype array (GH6471).Implemented
Panel.pct_change(GH6904)Added
howoption to rolling-moment functions to dictate how to handle resampling;rolling_max()defaults to max,rolling_min()defaults to min, and all others default to mean (GH6297)CustomBuisnessMonthBeginandCustomBusinessMonthEndare now available (GH6866)Series.quantile()andDataFrame.quantile()now accept an array of quantiles.describe()now accepts an array of percentiles to include in the summary statistics (GH4196)pivot_tablecan now acceptGrouperbyindexandcolumnskeywords (GH6913)In [72]: import datetime In [73]: df = DataFrame({ ....: 'Branch' : 'A A A A A B'.split(), ....: 'Buyer': 'Carl Mark Carl Carl Joe Joe'.split(), ....: 'Quantity': [1, 3, 5, 1, 8, 1], ....: 'Date' : [datetime.datetime(2013,11,1,13,0), datetime.datetime(2013,9,1,13,5), ....: datetime.datetime(2013,10,1,20,0), datetime.datetime(2013,10,2,10,0), ....: datetime.datetime(2013,11,1,20,0), datetime.datetime(2013,10,2,10,0)], ....: 'PayDay' : [datetime.datetime(2013,10,4,0,0), datetime.datetime(2013,10,15,13,5), ....: datetime.datetime(2013,9,5,20,0), datetime.datetime(2013,11,2,10,0), ....: datetime.datetime(2013,10,7,20,0), datetime.datetime(2013,9,5,10,0)]}) ....: In [74]: df Out[74]: Branch Buyer Date PayDay Quantity 0 A Carl 2013-11-01 13:00:00 2013-10-04 00:00:00 1 1 A Mark 2013-09-01 13:05:00 2013-10-15 13:05:00 3 2 A Carl 2013-10-01 20:00:00 2013-09-05 20:00:00 5 3 A Carl 2013-10-02 10:00:00 2013-11-02 10:00:00 1 4 A Joe 2013-11-01 20:00:00 2013-10-07 20:00:00 8 5 B Joe 2013-10-02 10:00:00 2013-09-05 10:00:00 1 In [75]: pivot_table(df, index=Grouper(freq='M', key='Date'), ....: columns=Grouper(freq='M', key='PayDay'), ....: values='Quantity', aggfunc=np.sum) ....: ���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[75]: PayDay 2013-09-30 2013-10-31 2013-11-30 Date 2013-09-30 NaN 3.0 NaN 2013-10-31 6.0 NaN 1.0 2013-11-30 NaN 9.0 NaN
Arrays of strings can be wrapped to a specified width (
str.wrap) (GH6999)Add
nsmallest()andSeries.nlargest()methods to Series, See the docs (GH3960)PeriodIndex fully supports partial string indexing like DatetimeIndex (GH7043)
In [76]: prng = period_range('2013-01-01 09:00', periods=100, freq='H') In [77]: ps = Series(np.random.randn(len(prng)), index=prng) In [78]: ps Out[78]: 2013-01-01 09:00 0.015696 2013-01-01 10:00 -2.242685 2013-01-01 11:00 1.150036 2013-01-01 12:00 0.991946 2013-01-01 13:00 0.953324 2013-01-01 14:00 -2.021255 2013-01-01 15:00 -0.334077 ... 2013-01-05 06:00 0.566534 2013-01-05 07:00 0.503592 2013-01-05 08:00 0.285296 2013-01-05 09:00 0.484288 2013-01-05 10:00 1.363482 2013-01-05 11:00 -0.781105 2013-01-05 12:00 -0.468018 Freq: H, Length: 100, dtype: float64 In [79]: ps['2013-01-02'] ��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[79]: 2013-01-02 00:00 0.553439 2013-01-02 01:00 1.318152 2013-01-02 02:00 -0.469305 2013-01-02 03:00 0.675554 2013-01-02 04:00 -1.817027 2013-01-02 05:00 -0.183109 2013-01-02 06:00 1.058969 ... 2013-01-02 17:00 0.076200 2013-01-02 18:00 -0.566446 2013-01-02 19:00 0.036142 2013-01-02 20:00 -2.074978 2013-01-02 21:00 0.247792 2013-01-02 22:00 -0.897157 2013-01-02 23:00 -0.136795 Freq: H, Length: 24, dtype: float64
read_excelcan now read milliseconds in Excel dates and times with xlrd >= 0.9.3. (GH5945)pd.stats.moments.rolling_varnow uses Welford’s method for increased numerical stability (GH6817)pd.expanding_apply and pd.rolling_apply now take args and kwargs that are passed on to the func (GH6289)
DataFrame.rank()now has a percentage rank option (GH5971)Series.rank()now has a percentage rank option (GH5971)Series.rank()andDataFrame.rank()now acceptmethod='dense'for ranks without gaps (GH6514)Support passing
encodingwith xlwt (GH3710)Refactor Block classes removing Block.items attributes to avoid duplication in item handling (GH6745, GH6988).
Testing statements updated to use specialized asserts (GH6175)
Performance¶
- Performance improvement when converting
DatetimeIndexto floating ordinals usingDatetimeConverter(GH6636) - Performance improvement for
DataFrame.shift(GH5609) - Performance improvement in indexing into a multi-indexed Series (GH5567)
- Performance improvements in single-dtyped indexing (GH6484)
- Improve performance of DataFrame construction with certain offsets, by removing faulty caching (e.g. MonthEnd,BusinessMonthEnd), (GH6479)
- Improve performance of
CustomBusinessDay(GH6584) - improve performance of slice indexing on Series with string keys (GH6341, GH6372)
- Performance improvement for
DataFrame.from_recordswhen reading a specified number of rows from an iterable (GH6700) - Performance improvements in timedelta conversions for integer dtypes (GH6754)
- Improved performance of compatible pickles (GH6899)
- Improve performance in certain reindexing operations by optimizing
take_2d(GH6749) GroupBy.count()is now implemented in Cython and is much faster for large numbers of groups (GH7016).
Experimental¶
There are no experimental changes in 0.14.0
Bug Fixes¶
- Bug in Series ValueError when index doesn’t match data (GH6532)
- Prevent segfault due to MultiIndex not being supported in HDFStore table format (GH1848)
- Bug in
pd.DataFrame.sort_indexwhere mergesort wasn’t stable whenascending=False(GH6399) - Bug in
pd.tseries.frequencies.to_offsetwhen argument has leading zeroes (GH6391) - Bug in version string gen. for dev versions with shallow clones / install from tarball (GH6127)
- Inconsistent tz parsing
Timestamp/to_datetimefor current year (GH5958) - Indexing bugs with reordered indexes (GH6252, GH6254)
- Bug in
.xswith a Series multiindex (GH6258, GH5684) - Bug in conversion of a string types to a DatetimeIndex with a specified frequency (GH6273, GH6274)
- Bug in
evalwhere type-promotion failed for large expressions (GH6205) - Bug in interpolate with
inplace=True(GH6281) HDFStore.removenow handles start and stop (GH6177)HDFStore.select_as_multiplehandles start and stop the same way asselect(GH6177)HDFStore.select_as_coordinatesandselect_columnworks with awhereclause that results in filters (GH6177)- Regression in join of non_unique_indexes (GH6329)
- Issue with groupby
aggwith a single function and a a mixed-type frame (GH6337) - Bug in
DataFrame.replace()when passing a non-boolto_replaceargument (GH6332) - Raise when trying to align on different levels of a multi-index assignment (GH3738)
- Bug in setting complex dtypes via boolean indexing (GH6345)
- Bug in TimeGrouper/resample when presented with a non-monotonic DatetimeIndex that would return invalid results. (GH4161)
- Bug in index name propogation in TimeGrouper/resample (GH4161)
- TimeGrouper has a more compatible API to the rest of the groupers (e.g.
groupswas missing) (GH3881) - Bug in multiple grouping with a TimeGrouper depending on target column order (GH6764)
- Bug in
pd.evalwhen parsing strings with possible tokens like'&'(GH6351) - Bug correctly handle placements of
-infin Panels when dividing by integer 0 (GH6178) DataFrame.shiftwithaxis=1was raising (GH6371)- Disabled clipboard tests until release time (run locally with
nosetests -A disabled) (GH6048). - Bug in
DataFrame.replace()when passing a nesteddictthat contained keys not in the values to be replaced (GH6342) str.matchignored the na flag (GH6609).- Bug in take with duplicate columns that were not consolidated (GH6240)
- Bug in interpolate changing dtypes (GH6290)
- Bug in
Series.get, was using a buggy access method (GH6383) - Bug in hdfstore queries of the form
where=[('date', '>=', datetime(2013,1,1)), ('date', '<=', datetime(2014,1,1))](GH6313) - Bug in
DataFrame.dropnawith duplicate indices (GH6355) - Regression in chained getitem indexing with embedded list-like from 0.12 (GH6394)
Float64Indexwith nans not comparing correctly (GH6401)eval/queryexpressions with strings containing the@character will now work (GH6366).- Bug in
Series.reindexwhen specifying amethodwith some nan values was inconsistent (noted on a resample) (GH6418) - Bug in
DataFrame.replace()where nested dicts were erroneously depending on the order of dictionary keys and values (GH5338). - Perf issue in concatting with empty objects (GH3259)
- Clarify sorting of
sym_diffonIndexobjects withNaNvalues (GH6444) - Regression in
MultiIndex.from_productwith aDatetimeIndexas input (GH6439) - Bug in
str.extractwhen passed a non-default index (GH6348) - Bug in
str.splitwhen passedpat=Noneandn=1(GH6466) - Bug in
io.data.DataReaderwhen passed"F-F_Momentum_Factor"anddata_source="famafrench"(GH6460) - Bug in
sumof atimedelta64[ns]series (GH6462) - Bug in
resamplewith a timezone and certain offsets (GH6397) - Bug in
iat/ilocwith duplicate indices on a Series (GH6493) - Bug in
read_htmlwhere nan’s were incorrectly being used to indicate missing values in text. Should use the empty string for consistency with the rest of pandas (GH5129). - Bug in
read_htmltests where redirected invalid URLs would make one test fail (GH6445). - Bug in multi-axis indexing using
.locon non-unique indices (GH6504) - Bug that caused _ref_locs corruption when slice indexing across columns axis of a DataFrame (GH6525)
- Regression from 0.13 in the treatment of numpy
datetime64non-ns dtypes in Series creation (GH6529) .namesattribute of MultiIndexes passed toset_indexare now preserved (GH6459).- Bug in setitem with a duplicate index and an alignable rhs (GH6541)
- Bug in setitem with
.locon mixed integer Indexes (GH6546) - Bug in
pd.read_statawhich would use the wrong data types and missing values (GH6327) - Bug in
DataFrame.to_statathat lead to data loss in certain cases, and could be exported using the wrong data types and missing values (GH6335) StataWriterreplaces missing values in string columns by empty string (GH6802)- Inconsistent types in
Timestampaddition/subtraction (GH6543) - Bug in preserving frequency across Timestamp addition/subtraction (GH4547)
- Bug in empty list lookup caused
IndexErrorexceptions (GH6536, GH6551) Series.quantileraising on anobjectdtype (GH6555)- Bug in
.xswith ananin level when dropped (GH6574) - Bug in fillna with
method='bfill/ffill'anddatetime64[ns]dtype (GH6587) - Bug in sql writing with mixed dtypes possibly leading to data loss (GH6509)
- Bug in
Series.pop(GH6600) - Bug in
ilocindexing when positional indexer matchedInt64Indexof the corresponding axis and no reordering happened (GH6612) - Bug in
fillnawithlimitandvaluespecified - Bug in
DataFrame.to_statawhen columns have non-string names (GH4558) - Bug in compat with
np.compress, surfaced in (GH6658) - Bug in binary operations with a rhs of a Series not aligning (GH6681)
- Bug in
DataFrame.to_statawhich incorrectly handles nan values and ignoreswith_indexkeyword argument (GH6685) - Bug in resample with extra bins when using an evenly divisible frequency (GH4076)
- Bug in consistency of groupby aggregation when passing a custom function (GH6715)
- Bug in resample when
how=Noneresample freq is the same as the axis frequency (GH5955) - Bug in downcasting inference with empty arrays (GH6733)
- Bug in
obj.blockson sparse containers dropping all but the last items of same for dtype (GH6748) - Bug in unpickling
NaT (NaTType)(GH4606) - Bug in
DataFrame.replace()where regex metacharacters were being treated as regexs even whenregex=False(GH6777). - Bug in timedelta ops on 32-bit platforms (GH6808)
- Bug in setting a tz-aware index directly via
.index(GH6785) - Bug in expressions.py where numexpr would try to evaluate arithmetic ops (GH6762).
- Bug in Makefile where it didn’t remove Cython generated C files with
make clean(GH6768) - Bug with numpy < 1.7.2 when reading long strings from
HDFStore(GH6166) - Bug in
DataFrame._reducewhere non bool-like (0/1) integers were being coverted into bools. (GH6806) - Regression from 0.13 with
fillnaand a Series on datetime-like (GH6344) - Bug in adding
np.timedelta64toDatetimeIndexwith timezone outputs incorrect results (GH6818) - Bug in
DataFrame.replace()where changing a dtype through replacement would only replace the first occurrence of a value (GH6689) - Better error message when passing a frequency of ‘MS’ in
Periodconstruction (GH5332) - Bug in
Series.__unicode__whenmax_rows=Noneand the Series has more than 1000 rows. (GH6863) - Bug in
groupby.get_groupwhere a datetlike wasn’t always accepted (GH5267) - Bug in
groupBy.get_groupcreated byTimeGrouperraisesAttributeError(GH6914) - Bug in
DatetimeIndex.tz_localizeandDatetimeIndex.tz_convertconvertingNaTincorrectly (GH5546) - Bug in arithmetic operations affecting
NaT(GH6873) - Bug in
Series.str.extractwhere the resultingSeriesfrom a single group match wasn’t renamed to the group name - Bug in
DataFrame.to_csvwhere settingindex=Falseignored theheaderkwarg (GH6186) - Bug in
DataFrame.plotandSeries.plot, where the legend behave inconsistently when plotting to the same axes repeatedly (GH6678) - Internal tests for patching
__finalize__/ bug in merge not finalizing (GH6923, GH6927) - accept
TextFileReaderinconcat, which was affecting a common user idiom (GH6583) - Bug in C parser with leading whitespace (GH3374)
- Bug in C parser with
delim_whitespace=Trueand\r-delimited lines - Bug in python parser with explicit multi-index in row following column header (GH6893)
- Bug in
Series.rankandDataFrame.rankthat caused small floats (<1e-13) to all receive the same rank (GH6886) - Bug in
DataFrame.applywith functions that used *args`` or **kwargs and returned an empty result (GH6952) - Bug in sum/mean on 32-bit platforms on overflows (GH6915)
- Moved
Panel.shifttoNDFrame.slice_shiftand fixed to respect multiple dtypes. (GH6959) - Bug in enabling
subplots=TrueinDataFrame.plotonly has single column raisesTypeError, andSeries.plotraisesAttributeError(GH6951) - Bug in
DataFrame.plotdraws unnecessary axes when enablingsubplotsandkind=scatter(GH6951) - Bug in
read_csvfrom a filesystem with non-utf-8 encoding (GH6807) - Bug in
ilocwhen setting / aligning (GH6766) - Bug causing UnicodeEncodeError when get_dummies called with unicode values and a prefix (GH6885)
- Bug in timeseries-with-frequency plot cursor display (GH5453)
- Bug surfaced in
groupby.plotwhen using aFloat64Index(GH7025) - Stopped tests from failing if options data isn’t able to be downloaded from Yahoo (GH7034)
- Bug in
parallel_coordinatesandradvizwhere reordering of class column caused possible color/class mismatch (GH6956) - Bug in
radvizandandrews_curveswhere multiple values of ‘color’ were being passed to plotting method (GH6956) - Bug in
Float64Index.isin()where containingnans would make indices claim that they contained all the things (GH7066). - Bug in
DataFrame.boxplotwhere it failed to use the axis passed as theaxargument (GH3578) - Bug in the
XlsxWriterandXlwtWriterimplementations that resulted in datetime columns being formatted without the time (GH7075) were being passed to plotting method read_fwf()treatsNoneincolspeclike regular python slices. It now reads from the beginning or until the end of the line whencolspeccontains aNone(previously raised aTypeError)- Bug in cache coherence with chained indexing and slicing; add
_is_viewproperty toNDFrameto correctly predict views; markis_copyonxsonly if its an actual copy (and not a view) (GH7084) - Bug in DatetimeIndex creation from string ndarray with
dayfirst=True(GH5917) - Bug in
MultiIndex.from_arrayscreated fromDatetimeIndexdoesn’t preservefreqandtz(GH7090) - Bug in
unstackraisesValueErrorwhenMultiIndexcontainsPeriodIndex(GH4342) - Bug in
boxplotandhistdraws unnecessary axes (GH6769) - Regression in
groupby.nth()for out-of-bounds indexers (GH6621) - Bug in
quantilewith datetime values (GH6965) - Bug in
Dataframe.set_index,reindexandpivotdon’t preserveDatetimeIndexandPeriodIndexattributes (GH3950, GH5878, GH6631) - Bug in
MultiIndex.get_level_valuesdoesn’t preserveDatetimeIndexandPeriodIndexattributes (GH7092) - Bug in
Groupbydoesn’t preservetz(GH3950) - Bug in
PeriodIndexpartial string slicing (GH6716) - Bug in the HTML repr of a truncated Series or DataFrame not showing the class name with the large_repr set to ‘info’ (GH7105)
- Bug in
DatetimeIndexspecifyingfreqraisesValueErrorwhen passed value is too short (GH7098) - Fixed a bug with the info repr not honoring the display.max_info_columns setting (GH6939)
- Bug
PeriodIndexstring slicing with out of bounds values (GH5407) - Fixed a memory error in the hashtable implementation/factorizer on resizing of large tables (GH7157)
- Bug in
isnullwhen applied to 0-dimensional object arrays (GH7176) - Bug in
query/evalwhere global constants were not looked up correctly (GH7178) - Bug in recognizing out-of-bounds positional list indexers with
ilocand a multi-axis tuple indexer (GH7189) - Bug in setitem with a single value, multi-index and integer indices (GH7190, GH7218)
- Bug in expressions evaluation with reversed ops, showing in series-dataframe ops (GH7198, GH7192)
- Bug in multi-axis indexing with > 2 ndim and a multi-index (GH7199)
- Fix a bug where invalid eval/query operations would blow the stack (GH5198)
v0.13.1 (February 3, 2014)¶
This is a minor release from 0.13.0 and includes a small number of API changes, several new features, enhancements, and performance improvements along with a large number of bug fixes. We recommend that all users upgrade to this version.
Highlights include:
- Added
infer_datetime_formatkeyword toread_csv/to_datetimeto allow speedups for homogeneously formatted datetimes. - Will intelligently limit display precision for datetime/timedelta formats.
- Enhanced Panel
apply()method. - Suggested tutorials in new Tutorials section.
- Our pandas ecosystem is growing, We now feature related projects in a new Pandas Ecosystem section.
- Much work has been taking place on improving the docs, and a new Contributing section has been added.
- Even though it may only be of interest to devs, we <3 our new CI status page: ScatterCI.
Warning
0.13.1 fixes a bug that was caused by a combination of having numpy < 1.8, and doing chained assignment on a string-like array. Please review the docs, chained indexing can have unexpected results and should generally be avoided.
This would previously segfault:
In [1]: df = DataFrame(dict(A = np.array(['foo','bar','bah','foo','bar'])))
In [2]: df['A'].iloc[0] = np.nan
In [3]: df
Out[3]:
A
0 NaN
1 bar
2 bah
3 foo
4 bar
The recommended way to do this type of assignment is:
In [4]: df = DataFrame(dict(A = np.array(['foo','bar','bah','foo','bar'])))
In [5]: df.loc[0,'A'] = np.nan
In [6]: df
Out[6]:
A
0 NaN
1 bar
2 bah
3 foo
4 bar
Output Formatting Enhancements¶
df.info() view now display dtype info per column (GH5682)
df.info() now honors the option
max_info_rows, to disable null counts for large frames (GH5974)In [7]: max_info_rows = pd.get_option('max_info_rows') In [8]: df = DataFrame(dict(A = np.random.randn(10), ...: B = np.random.randn(10), ...: C = date_range('20130101',periods=10))) ...: In [9]: df.iloc[3:6,[0,2]] = np.nan
# set to not display the null counts In [10]: pd.set_option('max_info_rows',0) In [11]: df.info() <class 'pandas.core.frame.DataFrame'> RangeIndex: 10 entries, 0 to 9 Data columns (total 3 columns): A float64 B float64 C datetime64[ns] dtypes: datetime64[ns](1), float64(2) memory usage: 320.0 bytes
# this is the default (same as in 0.13.0) In [12]: pd.set_option('max_info_rows',max_info_rows) In [13]: df.info() <class 'pandas.core.frame.DataFrame'> RangeIndex: 10 entries, 0 to 9 Data columns (total 3 columns): A 7 non-null float64 B 10 non-null float64 C 7 non-null datetime64[ns] dtypes: datetime64[ns](1), float64(2) memory usage: 320.0 bytes
Add
show_dimensionsdisplay option for the new DataFrame repr to control whether the dimensions print.In [14]: df = DataFrame([[1, 2], [3, 4]]) In [15]: pd.set_option('show_dimensions', False) In [16]: df Out[16]: 0 1 0 1 2 1 3 4 In [17]: pd.set_option('show_dimensions', True) In [18]: df Out[18]: 0 1 0 1 2 1 3 4 [2 rows x 2 columns]
The
ArrayFormatterfordatetimeandtimedelta64now intelligently limit precision based on the values in the array (GH3401)Previously output might look like:
age today diff 0 2001-01-01 00:00:00 2013-04-19 00:00:00 4491 days, 00:00:00 1 2004-06-01 00:00:00 2013-04-19 00:00:00 3244 days, 00:00:00
Now the output looks like:
In [19]: df = DataFrame([ Timestamp('20010101'), ....: Timestamp('20040601') ], columns=['age']) ....: In [20]: df['today'] = Timestamp('20130419') In [21]: df['diff'] = df['today']-df['age'] In [22]: df Out[22]: age today diff 0 2001-01-01 2013-04-19 4491 days 1 2004-06-01 2013-04-19 3244 days [2 rows x 3 columns]
API changes¶
Add
-NaNand-nanto the default set of NA values (GH5952). See NA Values.Added
Series.str.get_dummiesvectorized string method (GH6021), to extract dummy/indicator variables for separated string columns:In [23]: s = Series(['a', 'a|b', np.nan, 'a|c']) In [24]: s.str.get_dummies(sep='|') Out[24]: a b c 0 1 0 0 1 1 1 0 2 0 0 0 3 1 0 1 [4 rows x 3 columns]
Added the
NDFrame.equals()method to compare if two NDFrames are equal have equal axes, dtypes, and values. Added thearray_equivalentfunction to compare if two ndarrays are equal. NaNs in identical locations are treated as equal. (GH5283) See also the docs for a motivating example.In [25]: df = DataFrame({'col':['foo', 0, np.nan]}) In [26]: df2 = DataFrame({'col':[np.nan, 0, 'foo']}, index=[2,1,0]) In [27]: df.equals(df2) Out[27]: False In [28]: df.equals(df2.sort_index()) ���������������Out[28]: True In [29]: import pandas.core.common as com In [30]: com.array_equivalent(np.array([0, np.nan]), np.array([0, np.nan])) Out[30]: True In [31]: np.array_equal(np.array([0, np.nan]), np.array([0, np.nan])) ��������������Out[31]: False
DataFrame.applywill use thereduceargument to determine whether aSeriesor aDataFrameshould be returned when theDataFrameis empty (GH6007).Previously, calling
DataFrame.applyan emptyDataFramewould return either aDataFrameif there were no columns, or the function being applied would be called with an emptySeriesto guess whether aSeriesorDataFrameshould be returned:In [32]: def applied_func(col): ....: print("Apply function being called with: ", col) ....: return col.sum() ....: In [33]: empty = DataFrame(columns=['a', 'b']) In [34]: empty.apply(applied_func) Apply function being called with: Series([], Length: 0, dtype: float64) Out[34]: a NaN b NaN Length: 2, dtype: float64
Now, when
applyis called on an emptyDataFrame: if thereduceargument isTrueaSerieswill returned, if it isFalseaDataFramewill be returned, and if it isNone(the default) the function being applied will be called with an empty series to try and guess the return type.In [35]: empty.apply(applied_func, reduce=True) Out[35]: a NaN b NaN Length: 2, dtype: float64 In [36]: empty.apply(applied_func, reduce=False) ����������������������������������������������������Out[36]: Empty DataFrame Columns: [a, b] Index: [] [0 rows x 2 columns]
Prior Version Deprecations/Changes¶
There are no announced changes in 0.13 or prior that are taking effect as of 0.13.1
Deprecations¶
There are no deprecations of prior behavior in 0.13.1
Enhancements¶
pd.read_csvandpd.to_datetimelearned a newinfer_datetime_formatkeyword which greatly improves parsing perf in many cases. Thanks to @lexual for suggesting and @danbirken for rapidly implementing. (GH5490, GH6021)If
parse_datesis enabled and this flag is set, pandas will attempt to infer the format of the datetime strings in the columns, and if it can be inferred, switch to a faster method of parsing them. In some cases this can increase the parsing speed by ~5-10x.# Try to infer the format for the index column df = pd.read_csv('foo.csv', index_col=0, parse_dates=True, infer_datetime_format=True)
date_formatanddatetime_formatkeywords can now be specified when writing toexcelfiles (GH4133)MultiIndex.from_productconvenience function for creating a MultiIndex from the cartesian product of a set of iterables (GH6055):In [37]: shades = ['light', 'dark'] In [38]: colors = ['red', 'green', 'blue'] In [39]: MultiIndex.from_product([shades, colors], names=['shade', 'color']) Out[39]: MultiIndex(levels=[['dark', 'light'], ['blue', 'green', 'red']], labels=[[1, 1, 1, 0, 0, 0], [2, 1, 0, 2, 1, 0]], names=['shade', 'color'])
Panel
apply()will work on non-ufuncs. See the docs.In [40]: import pandas.util.testing as tm In [41]: panel = tm.makePanel(5) In [42]: panel Out[42]: <class 'pandas.core.panel.Panel'> Dimensions: 3 (items) x 5 (major_axis) x 4 (minor_axis) Items axis: ItemA to ItemC Major_axis axis: 2000-01-03 00:00:00 to 2000-01-07 00:00:00 Minor_axis axis: A to D In [43]: panel['ItemA'] �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[43]: A B C D 2000-01-03 0.694103 1.893534 -1.735349 -0.850346 2000-01-04 0.678630 0.639633 1.210384 1.176812 2000-01-05 0.239556 -0.962029 0.797435 -0.524336 2000-01-06 0.151227 -2.085266 -0.379811 0.700908 2000-01-07 0.816127 1.930247 0.702562 0.984188 [5 rows x 4 columns]
Specifying an
applythat operates on a Series (to return a single element)In [44]: panel.apply(lambda x: x.dtype, axis='items') Out[44]: A B C D 2000-01-03 float64 float64 float64 float64 2000-01-04 float64 float64 float64 float64 2000-01-05 float64 float64 float64 float64 2000-01-06 float64 float64 float64 float64 2000-01-07 float64 float64 float64 float64 [5 rows x 4 columns]
A similar reduction type operation
In [45]: panel.apply(lambda x: x.sum(), axis='major_axis') Out[45]: ItemA ItemB ItemC A 2.579643 3.062757 0.379252 B 1.416120 -1.960855 0.923558 C 0.595222 -1.079772 -3.118269 D 1.487226 -0.734611 -1.979310 [4 rows x 3 columns]
This is equivalent to
In [46]: panel.sum('major_axis') Out[46]: ItemA ItemB ItemC A 2.579643 3.062757 0.379252 B 1.416120 -1.960855 0.923558 C 0.595222 -1.079772 -3.118269 D 1.487226 -0.734611 -1.979310 [4 rows x 3 columns]
A transformation operation that returns a Panel, but is computing the z-score across the major_axis
In [47]: result = panel.apply( ....: lambda x: (x-x.mean())/x.std(), ....: axis='major_axis') ....: In [48]: result Out[48]: <class 'pandas.core.panel.Panel'> Dimensions: 3 (items) x 5 (major_axis) x 4 (minor_axis) Items axis: ItemA to ItemC Major_axis axis: 2000-01-03 00:00:00 to 2000-01-07 00:00:00 Minor_axis axis: A to D In [49]: result['ItemA'] �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[49]: A B C D 2000-01-03 0.595800 0.907552 -1.556260 -1.244875 2000-01-04 0.544058 0.200868 0.915883 0.953747 2000-01-05 -0.924165 -0.701810 0.569325 -0.891290 2000-01-06 -1.219530 -1.334852 -0.418654 0.437589 2000-01-07 1.003837 0.928242 0.489705 0.744830 [5 rows x 4 columns]
Panel
apply()operating on cross-sectional slabs. (GH1148)In [50]: f = lambda x: ((x.T-x.mean(1))/x.std(1)).T In [51]: result = panel.apply(f, axis = ['items','major_axis']) In [52]: result Out[52]: <class 'pandas.core.panel.Panel'> Dimensions: 4 (items) x 5 (major_axis) x 3 (minor_axis) Items axis: A to D Major_axis axis: 2000-01-03 00:00:00 to 2000-01-07 00:00:00 Minor_axis axis: ItemA to ItemC In [53]: result.loc[:,:,'ItemA'] �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[53]: A B C D 2000-01-03 0.331409 1.071034 -0.914540 -0.510587 2000-01-04 -0.741017 -0.118794 0.383277 0.537212 2000-01-05 0.065042 -0.767353 0.655436 0.069467 2000-01-06 0.027932 -0.569477 0.908202 0.610585 2000-01-07 1.116434 1.133591 0.871287 1.004064 [5 rows x 4 columns]
This is equivalent to the following
In [54]: result = Panel(dict([ (ax,f(panel.loc[:,:,ax])) ....: for ax in panel.minor_axis ])) ....: In [55]: result Out[55]: <class 'pandas.core.panel.Panel'> Dimensions: 4 (items) x 5 (major_axis) x 3 (minor_axis) Items axis: A to D Major_axis axis: 2000-01-03 00:00:00 to 2000-01-07 00:00:00 Minor_axis axis: ItemA to ItemC In [56]: result.loc[:,:,'ItemA'] �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[56]: A B C D 2000-01-03 0.331409 1.071034 -0.914540 -0.510587 2000-01-04 -0.741017 -0.118794 0.383277 0.537212 2000-01-05 0.065042 -0.767353 0.655436 0.069467 2000-01-06 0.027932 -0.569477 0.908202 0.610585 2000-01-07 1.116434 1.133591 0.871287 1.004064 [5 rows x 4 columns]
Performance¶
Performance improvements for 0.13.1
- Series datetime/timedelta binary operations (GH5801)
- DataFrame
count/dropnaforaxis=1 - Series.str.contains now has a regex=False keyword which can be faster for plain (non-regex) string patterns. (GH5879)
- Series.str.extract (GH5944)
dtypes/ftypesmethods (GH5968)- indexing with object dtypes (GH5968)
DataFrame.apply(GH6013)- Regression in JSON IO (GH5765)
- Index construction from Series (GH6150)
Experimental¶
There are no experimental changes in 0.13.1
Bug Fixes¶
See V0.13.1 Bug Fixes for an extensive list of bugs that have been fixed in 0.13.1.
See the full release notes or issue tracker on GitHub for a complete list of all API changes, Enhancements and Bug Fixes.
v0.13.0 (January 3, 2014)¶
This is a major release from 0.12.0 and includes a number of API changes, several new features and enhancements along with a large number of bug fixes.
Highlights include:
- support for a new index type
Float64Index, and other Indexing enhancements HDFStorehas a new string based syntax for query specification- support for new methods of interpolation
- updated
timedeltaoperations - a new string manipulation method
extract - Nanosecond support for Offsets
isinfor DataFrames
Several experimental features are added, including:
- new
eval/querymethods for expression evaluation - support for
msgpackserialization - an i/o interface to Google’s
BigQuery
Their are several new or updated docs sections including:
- Comparison with SQL, which should be useful for those familiar with SQL but still learning pandas.
- Comparison with R, idiom translations from R to pandas.
- Enhancing Performance, ways to enhance pandas performance with
eval/query.
Warning
In 0.13.0 Series has internally been refactored to no longer sub-class ndarray
but instead subclass NDFrame, similar to the rest of the pandas containers. This should be
a transparent change with only very limited API implications. See Internal Refactoring
API changes¶
read_excelnow supports an integer in itssheetnameargument giving the index of the sheet to read in (GH4301).Text parser now treats anything that reads like inf (“inf”, “Inf”, “-Inf”, “iNf”, etc.) as infinity. (GH4220, GH4219), affecting
read_table,read_csv, etc.pandasnow is Python 2/3 compatible without the need for 2to3 thanks to @jtratner. As a result, pandas now uses iterators more extensively. This also led to the introduction of substantive parts of the Benjamin Peterson’ssixlibrary into compat. (GH4384, GH4375, GH4372)pandas.util.compatandpandas.util.py3compathave been merged intopandas.compat.pandas.compatnow includes many functions allowing 2/3 compatibility. It contains both list and iterator versions of range, filter, map and zip, plus other necessary elements for Python 3 compatibility.lmap,lzip,lrangeandlfilterall produce lists instead of iterators, for compatibility withnumpy, subscripting andpandasconstructors.(GH4384, GH4375, GH4372)Series.getwith negative indexers now returns the same as[](GH4390)Changes to how
IndexandMultiIndexhandle metadata (levels,labels, andnames) (GH4039):# previously, you would have set levels or labels directly index.levels = [[1, 2, 3, 4], [1, 2, 4, 4]] # now, you use the set_levels or set_labels methods index = index.set_levels([[1, 2, 3, 4], [1, 2, 4, 4]]) # similarly, for names, you can rename the object # but setting names is not deprecated index = index.set_names(["bob", "cranberry"]) # and all methods take an inplace kwarg - but return None index.set_names(["bob", "cranberry"], inplace=True)
All division with
NDFrameobjects is now truedivision, regardless of the future import. This means that operating on pandas objects will by default use floating point division, and return a floating point dtype. You can use//andfloordivto do integer division.Integer division
In [3]: arr = np.array([1, 2, 3, 4]) In [4]: arr2 = np.array([5, 3, 2, 1]) In [5]: arr / arr2 Out[5]: array([0, 0, 1, 4]) In [6]: Series(arr) // Series(arr2) Out[6]: 0 0 1 0 2 1 3 4 dtype: int64
True Division
In [7]: pd.Series(arr) / pd.Series(arr2) # no future import required Out[7]: 0 0.200000 1 0.666667 2 1.500000 3 4.000000 dtype: float64
Infer and downcast dtype if
downcast='infer'is passed tofillna/ffill/bfill(GH4604)__nonzero__for all NDFrame objects, will now raise aValueError, this reverts back to (GH1073, GH4633) behavior. See gotchas for a more detailed discussion.This prevents doing boolean comparison on entire pandas objects, which is inherently ambiguous. These all will raise a
ValueError.if df: .... df1 and df2 s1 and s2
Added the
.bool()method toNDFrameobjects to facilitate evaluating of single-element boolean Series:In [1]: Series([True]).bool() Out[1]: True In [2]: Series([False]).bool() �������������Out[2]: False In [3]: DataFrame([[True]]).bool() ���������������������������Out[3]: True In [4]: DataFrame([[False]]).bool() ����������������������������������������Out[4]: False
All non-Index NDFrames (
Series,DataFrame,Panel,Panel4D,SparsePanel, etc.), now support the entire set of arithmetic operators and arithmetic flex methods (add, sub, mul, etc.).SparsePaneldoes not supportpowormodwith non-scalars. (GH3765)SeriesandDataFramenow have amode()method to calculate the statistical mode(s) by axis/Series. (GH5367)Chained assignment will now by default warn if the user is assigning to a copy. This can be changed with the option
mode.chained_assignment, allowed options areraise/warn/None. See the docs.In [5]: dfc = DataFrame({'A':['aaa','bbb','ccc'],'B':[1,2,3]}) In [6]: pd.set_option('chained_assignment','warn')
The following warning / exception will show if this is attempted.
In [7]: dfc.loc[0]['A'] = 1111
Traceback (most recent call last) ... SettingWithCopyWarning: A value is trying to be set on a copy of a slice from a DataFrame. Try using .loc[row_index,col_indexer] = value instead
Here is the correct method of assignment.
In [8]: dfc.loc[0,'A'] = 11 In [9]: dfc Out[9]: A B 0 11 1 1 bbb 2 2 ccc 3 [3 rows x 2 columns]
Panel.reindexhas the following call signaturePanel.reindex(items=None, major_axis=None, minor_axis=None, **kwargs)to conform with other
NDFrameobjects. See Internal Refactoring for more information.
Series.argminandSeries.argmaxare now aliased toSeries.idxminandSeries.idxmax. These return the index of themin or max element respectively. Prior to 0.13.0 these would return the position of the min / max element. (GH6214)
Prior Version Deprecations/Changes¶
These were announced changes in 0.12 or prior that are taking effect as of 0.13.0
- Remove deprecated
Factor(GH3650) - Remove deprecated
set_printoptions/reset_printoptions(GH3046) - Remove deprecated
_verbose_info(GH3215) - Remove deprecated
read_clipboard/to_clipboard/ExcelFile/ExcelWriterfrompandas.io.parsers(GH3717) These are available as functions in the main pandas namespace (e.g.pd.read_clipboard) - default for
tupleize_colsis nowFalsefor bothto_csvandread_csv. Fair warning in 0.12 (GH3604) - default for display.max_seq_len is now 100 rather then None. This activates truncated display (”...”) of long sequences in various places. (GH3391)
Deprecations¶
Deprecated in 0.13.0
- deprecated
iterkv, which will be removed in a future release (this was an alias of iteritems used to bypass2to3‘s changes). (GH4384, GH4375, GH4372) - deprecated the string method
match, whose role is now performed more idiomatically byextract. In a future release, the default behavior ofmatchwill change to become analogous tocontains, which returns a boolean indexer. (Their distinction is strictness:matchrelies onre.matchwhilecontainsrelies onre.search.) In this release, the deprecated behavior is the default, but the new behavior is available through the keyword argumentas_indexer=True.
Indexing API Changes¶
Prior to 0.13, it was impossible to use a label indexer (.loc/.ix) to set a value that
was not contained in the index of a particular axis. (GH2578). See the docs
In the Series case this is effectively an appending operation
In [10]: s = Series([1,2,3])
In [11]: s
Out[11]:
0 1
1 2
2 3
Length: 3, dtype: int64
In [12]: s[5] = 5.
In [13]: s
Out[13]:
0 1.0
1 2.0
2 3.0
5 5.0
Length: 4, dtype: float64
In [14]: dfi = DataFrame(np.arange(6).reshape(3,2),
....: columns=['A','B'])
....:
In [15]: dfi
Out[15]:
A B
0 0 1
1 2 3
2 4 5
[3 rows x 2 columns]
This would previously KeyError
In [16]: dfi.loc[:,'C'] = dfi.loc[:,'A']
In [17]: dfi
Out[17]:
A B C
0 0 1 0
1 2 3 2
2 4 5 4
[3 rows x 3 columns]
This is like an append operation.
In [18]: dfi.loc[3] = 5
In [19]: dfi
Out[19]:
A B C
0 0 1 0
1 2 3 2
2 4 5 4
3 5 5 5
[4 rows x 3 columns]
A Panel setting operation on an arbitrary axis aligns the input to the Panel
In [20]: p = pd.Panel(np.arange(16).reshape(2,4,2),
....: items=['Item1','Item2'],
....: major_axis=pd.date_range('2001/1/12',periods=4),
....: minor_axis=['A','B'],dtype='float64')
....:
In [21]: p
Out[21]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 4 (major_axis) x 2 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 2001-01-12 00:00:00 to 2001-01-15 00:00:00
Minor_axis axis: A to B
In [22]: p.loc[:,:,'C'] = Series([30,32],index=p.items)
In [23]: p
Out[23]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 4 (major_axis) x 3 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 2001-01-12 00:00:00 to 2001-01-15 00:00:00
Minor_axis axis: A to C
In [24]: p.loc[:,:,'C']
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[24]:
Item1 Item2
2001-01-12 30.0 32.0
2001-01-13 30.0 32.0
2001-01-14 30.0 32.0
2001-01-15 30.0 32.0
[4 rows x 2 columns]
Float64Index API Change¶
Added a new index type,
Float64Index. This will be automatically created when passing floating values in index creation. This enables a pure label-based slicing paradigm that makes[],ix,locfor scalar indexing and slicing work exactly the same. See the docs, (GH263)Construction is by default for floating type values.
In [25]: index = Index([1.5, 2, 3, 4.5, 5]) In [26]: index Out[26]: Float64Index([1.5, 2.0, 3.0, 4.5, 5.0], dtype='float64') In [27]: s = Series(range(5),index=index) In [28]: s Out[28]: 1.5 0 2.0 1 3.0 2 4.5 3 5.0 4 Length: 5, dtype: int64
Scalar selection for
[],.ix,.locwill always be label based. An integer will match an equal float index (e.g.3is equivalent to3.0)In [29]: s[3] Out[29]: 2 In [30]: s.loc[3] �����������Out[30]: 2
The only positional indexing is via
ilocIn [31]: s.iloc[3] Out[31]: 3
A scalar index that is not found will raise
KeyErrorSlicing is ALWAYS on the values of the index, for
[],ix,locand ALWAYS positional withilocIn [32]: s[2:4] Out[32]: 2.0 1 3.0 2 Length: 2, dtype: int64 In [33]: s.loc[2:4] ����������������������������������������������������Out[33]: 2.0 1 3.0 2 Length: 2, dtype: int64 In [34]: s.iloc[2:4] ��������������������������������������������������������������������������������������������������������Out[34]: 3.0 2 4.5 3 Length: 2, dtype: int64
In float indexes, slicing using floats are allowed
In [35]: s[2.1:4.6] Out[35]: 3.0 2 4.5 3 Length: 2, dtype: int64 In [36]: s.loc[2.1:4.6] ����������������������������������������������������Out[36]: 3.0 2 4.5 3 Length: 2, dtype: int64
Indexing on other index types are preserved (and positional fallback for
[],ix), with the exception, that floating point slicing on indexes on nonFloat64Indexwill now raise aTypeError.In [1]: Series(range(5))[3.5] TypeError: the label [3.5] is not a proper indexer for this index type (Int64Index) In [1]: Series(range(5))[3.5:4.5] TypeError: the slice start [3.5] is not a proper indexer for this index type (Int64Index)
Using a scalar float indexer will be deprecated in a future version, but is allowed for now.
In [3]: Series(range(5))[3.0] Out[3]: 3
HDFStore API Changes¶
Query Format Changes. A much more string-like query format is now supported. See the docs.
In [37]: path = 'test.h5' In [38]: dfq = DataFrame(randn(10,4), ....: columns=list('ABCD'), ....: index=date_range('20130101',periods=10)) ....: In [39]: dfq.to_hdf(path,'dfq',format='table',data_columns=True)
Use boolean expressions, with in-line function evaluation.
In [40]: read_hdf(path,'dfq', ....: where="index>Timestamp('20130104') & columns=['A', 'B']") ....: Out[40]: A B 2013-01-05 1.057633 -0.791489 2013-01-06 1.910759 0.787965 2013-01-07 1.043945 2.107785 2013-01-08 0.749185 -0.675521 2013-01-09 -0.276646 1.924533 2013-01-10 0.226363 -2.078618 [6 rows x 2 columns]
Use an inline column reference
In [41]: read_hdf(path,'dfq', ....: where="A>0 or C>0") ....: Out[41]: A B C D 2013-01-01 -0.414505 -1.425795 0.209395 -0.592886 2013-01-02 -1.473116 -0.896581 1.104352 -0.431550 2013-01-03 -0.161137 0.889157 0.288377 -1.051539 2013-01-04 -0.319561 -0.619993 0.156998 -0.571455 2013-01-05 1.057633 -0.791489 -0.524627 0.071878 2013-01-06 1.910759 0.787965 0.513082 -0.546416 2013-01-07 1.043945 2.107785 1.459927 1.015405 2013-01-08 0.749185 -0.675521 0.440266 0.688972 2013-01-09 -0.276646 1.924533 0.411204 0.890765 2013-01-10 0.226363 -2.078618 -0.387886 -0.087107 [10 rows x 4 columns]
the
formatkeyword now replaces thetablekeyword; allowed values arefixed(f)ortable(t)the same defaults as prior < 0.13.0 remain, e.g.putimpliesfixedformat andappendimpliestableformat. This default format can be set as an option by settingio.hdf.default_format.In [42]: path = 'test.h5' In [43]: df = pd.DataFrame(np.random.randn(10,2)) In [44]: df.to_hdf(path,'df_table',format='table') In [45]: df.to_hdf(path,'df_table2',append=True) In [46]: df.to_hdf(path,'df_fixed') In [47]: with pd.HDFStore(path) as store: ....: print(store) ....: <class 'pandas.io.pytables.HDFStore'> File path: test.h5 /df_fixed frame (shape->[10,2]) /df_table frame_table (typ->appendable,nrows->10,ncols->2,indexers->[index]) /df_table2 frame_table (typ->appendable,nrows->10,ncols->2,indexers->[index])
Significant table writing performance improvements
handle a passed
Seriesin table format (GH4330)can now serialize a
timedelta64[ns]dtype in a table (GH3577), See the docs.added an
is_openproperty to indicate if the underlying file handle is_open; a closed store will now report ‘CLOSED’ when viewing the store (rather than raising an error) (GH4409)a close of a
HDFStorenow will close that instance of theHDFStorebut will only close the actual file if the ref count (byPyTables) w.r.t. all of the open handles are 0. Essentially you have a local instance ofHDFStorereferenced by a variable. Once you close it, it will report closed. Other references (to the same file) will continue to operate until they themselves are closed. Performing an action on a closed file will raiseClosedFileErrorIn [48]: path = 'test.h5' In [49]: df = DataFrame(randn(10,2)) In [50]: store1 = HDFStore(path) In [51]: store2 = HDFStore(path) In [52]: store1.append('df',df) In [53]: store2.append('df2',df) In [54]: store1 Out[54]: <class 'pandas.io.pytables.HDFStore'> File path: test.h5 /df frame_table (typ->appendable,nrows->10,ncols->2,indexers->[index]) In [55]: store2 ������������������������������������������������������������������������������������������������������������������������������������������������������Out[55]: <class 'pandas.io.pytables.HDFStore'> File path: test.h5 /df frame_table (typ->appendable,nrows->10,ncols->2,indexers->[index]) /df2 frame_table (typ->appendable,nrows->10,ncols->2,indexers->[index]) In [56]: store1.close() In [57]: store2 Out[57]: <class 'pandas.io.pytables.HDFStore'> File path: test.h5 /df frame_table (typ->appendable,nrows->10,ncols->2,indexers->[index]) /df2 frame_table (typ->appendable,nrows->10,ncols->2,indexers->[index]) In [58]: store2.close() In [59]: store2 Out[59]: <class 'pandas.io.pytables.HDFStore'> File path: test.h5 File is CLOSED
removed the
_quietattribute, replace by aDuplicateWarningif retrieving duplicate rows from a table (GH4367)removed the
warnargument fromopen. Instead aPossibleDataLossErrorexception will be raised if you try to usemode='w'with an OPEN file handle (GH4367)allow a passed locations array or mask as a
wherecondition (GH4467). See the docs for an example.add the keyword
dropna=Truetoappendto change whether ALL nan rows are not written to the store (default isTrue, ALL nan rows are NOT written), also settable via the optionio.hdf.dropna_table(GH4625)pass thru store creation arguments; can be used to support in-memory stores
DataFrame repr Changes¶
The HTML and plain text representations of DataFrame now show
a truncated view of the table once it exceeds a certain size, rather
than switching to the short info view (GH4886, GH5550).
This makes the representation more consistent as small DataFrames get
larger.
To get the info view, call DataFrame.info(). If you prefer the
info view as the repr for large DataFrames, you can set this by running
set_option('display.large_repr', 'info').
Enhancements¶
df.to_clipboard()learned a newexcelkeyword that let’s you paste df data directly into excel (enabled by default). (GH5070).read_htmlnow raises aURLErrorinstead of catching and raising aValueError(GH4303, GH4305)Added a test for
read_clipboard()andto_clipboard()(GH4282)Clipboard functionality now works with PySide (GH4282)
Added a more informative error message when plot arguments contain overlapping color and style arguments (GH4402)
to_dictnow takesrecordsas a possible outtype. Returns an array of column-keyed dictionaries. (GH4936)NaNhanding in get_dummies (GH4446) with dummy_na# previously, nan was erroneously counted as 2 here # now it is not counted at all In [60]: get_dummies([1, 2, np.nan]) Out[60]: 1.0 2.0 0 1 0 1 0 1 2 0 0 [3 rows x 2 columns] # unless requested In [61]: get_dummies([1, 2, np.nan], dummy_na=True) ��������������������������������������������������������������������������������Out[61]: 1.0 2.0 NaN 0 1 0 0 1 0 1 0 2 0 0 1 [3 rows x 3 columns]
timedelta64[ns]operations. See the docs.Warning
Most of these operations require
numpy >= 1.7Using the new top-level
to_timedelta, you can convert a scalar or array from the standard timedelta format (produced byto_csv) into a timedelta type (np.timedelta64innanoseconds).In [62]: to_timedelta('1 days 06:05:01.00003') Out[62]: Timedelta('1 days 06:05:01.000030') In [63]: to_timedelta('15.5us') ���������������������������������������������Out[63]: Timedelta('0 days 00:00:00.000015') In [64]: to_timedelta(['1 days 06:05:01.00003','15.5us','nan']) ������������������������������������������������������������������������������������������Out[64]: TimedeltaIndex(['1 days 06:05:01.000030', '0 days 00:00:00.000015', NaT], dtype='timedelta64[ns]', freq=None) In [65]: to_timedelta(np.arange(5),unit='s') �����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[65]: TimedeltaIndex(['00:00:00', '00:00:01', '00:00:02', '00:00:03', '00:00:04'], dtype='timedelta64[ns]', freq=None) In [66]: to_timedelta(np.arange(5),unit='d') �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[66]: TimedeltaIndex(['0 days', '1 days', '2 days', '3 days', '4 days'], dtype='timedelta64[ns]', freq=None)
A Series of dtype
timedelta64[ns]can now be divided by anothertimedelta64[ns]object, or astyped to yield afloat64dtyped Series. This is frequency conversion. See the docs for the docs.In [67]: from datetime import timedelta In [68]: td = Series(date_range('20130101',periods=4))-Series(date_range('20121201',periods=4)) In [69]: td[2] += np.timedelta64(timedelta(minutes=5,seconds=3)) In [70]: td[3] = np.nan In [71]: td Out[71]: 0 31 days 00:00:00 1 31 days 00:00:00 2 31 days 00:05:03 3 NaT Length: 4, dtype: timedelta64[ns] # to days In [72]: td / np.timedelta64(1,'D') ��������������������������������������������������������������������������������������������������������������������������������Out[72]: 0 31.000000 1 31.000000 2 31.003507 3 NaN Length: 4, dtype: float64 In [73]: td.astype('timedelta64[D]') ��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[73]: 0 31.0 1 31.0 2 31.0 3 NaN Length: 4, dtype: float64 # to seconds In [74]: td / np.timedelta64(1,'s') ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[74]: 0 2678400.0 1 2678400.0 2 2678703.0 3 NaN Length: 4, dtype: float64 In [75]: td.astype('timedelta64[s]') ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[75]: 0 2678400.0 1 2678400.0 2 2678703.0 3 NaN Length: 4, dtype: float64
Dividing or multiplying a
timedelta64[ns]Series by an integer or integer SeriesIn [76]: td * -1 Out[76]: 0 -31 days +00:00:00 1 -31 days +00:00:00 2 -32 days +23:54:57 3 NaT Length: 4, dtype: timedelta64[ns] In [77]: td * Series([1,2,3,4]) ����������������������������������������������������������������������������������������������������������������������������������������Out[77]: 0 31 days 00:00:00 1 62 days 00:00:00 2 93 days 00:15:09 3 NaT Length: 4, dtype: timedelta64[ns]
Absolute
DateOffsetobjects can act equivalently totimedeltasIn [78]: from pandas import offsets In [79]: td + offsets.Minute(5) + offsets.Milli(5) Out[79]: 0 31 days 00:05:00.005000 1 31 days 00:05:00.005000 2 31 days 00:10:03.005000 3 NaT Length: 4, dtype: timedelta64[ns]
Fillna is now supported for timedeltas
In [80]: td.fillna(0) Out[80]: 0 31 days 00:00:00 1 31 days 00:00:00 2 31 days 00:05:03 3 0 days 00:00:00 Length: 4, dtype: timedelta64[ns] In [81]: td.fillna(timedelta(days=1,seconds=5)) ��������������������������������������������������������������������������������������������������������������������������������Out[81]: 0 31 days 00:00:00 1 31 days 00:00:00 2 31 days 00:05:03 3 1 days 00:00:05 Length: 4, dtype: timedelta64[ns]
You can do numeric reduction operations on timedeltas.
In [82]: td.mean() Out[82]: Timedelta('31 days 00:01:41') In [83]: td.quantile(.1) ���������������������������������������Out[83]: Timedelta('31 days 00:00:00')
plot(kind='kde')now accepts the optional parametersbw_methodandind, passed to scipy.stats.gaussian_kde() (for scipy >= 0.11.0) to set the bandwidth, and to gkde.evaluate() to specify the indices at which it is evaluated, respectively. See scipy docs. (GH4298)DataFrame constructor now accepts a numpy masked record array (GH3478)
The new vectorized string method
extractreturn regular expression matches more conveniently.In [84]: Series(['a1', 'b2', 'c3']).str.extract('[ab](\d)') Out[84]: 0 1 1 2 2 NaN Length: 3, dtype: object
Elements that do not match return
NaN. Extracting a regular expression with more than one group returns a DataFrame with one column per group.In [85]: Series(['a1', 'b2', 'c3']).str.extract('([ab])(\d)') Out[85]: 0 1 0 a 1 1 b 2 2 NaN NaN [3 rows x 2 columns]
Elements that do not match return a row of
NaN. Thus, a Series of messy strings can be converted into a like-indexed Series or DataFrame of cleaned-up or more useful strings, without necessitatingget()to access tuples orre.matchobjects.Named groups like
In [86]: Series(['a1', 'b2', 'c3']).str.extract( ....: '(?P<letter>[ab])(?P<digit>\d)') ....: Out[86]: letter digit 0 a 1 1 b 2 2 NaN NaN [3 rows x 2 columns]
and optional groups can also be used.
In [87]: Series(['a1', 'b2', '3']).str.extract( ....: '(?P<letter>[ab])?(?P<digit>\d)') ....: Out[87]: letter digit 0 a 1 1 b 2 2 NaN 3 [3 rows x 2 columns]
read_statanow accepts Stata 13 format (GH4291)read_fwfnow infers the column specifications from the first 100 rows of the file if the data has correctly separated and properly aligned columns using the delimiter provided to the function (GH4488).support for nanosecond times as an offset
Warning
These operations require
numpy >= 1.7Period conversions in the range of seconds and below were reworked and extended up to nanoseconds. Periods in the nanosecond range are now available.
In [88]: date_range('2013-01-01', periods=5, freq='5N') Out[88]: DatetimeIndex(['2013-01-01', '2013-01-01', '2013-01-01', '2013-01-01', '2013-01-01'], dtype='datetime64[ns]', freq='5N')
or with frequency as offset
In [89]: date_range('2013-01-01', periods=5, freq=pd.offsets.Nano(5)) Out[89]: DatetimeIndex(['2013-01-01', '2013-01-01', '2013-01-01', '2013-01-01', '2013-01-01'], dtype='datetime64[ns]', freq='5N')
Timestamps can be modified in the nanosecond range
In [90]: t = Timestamp('20130101 09:01:02') In [91]: t + pd.tseries.offsets.Nano(123) Out[91]: Timestamp('2013-01-01 09:01:02.000000123')
A new method,
isinfor DataFrames, which plays nicely with boolean indexing. The argument toisin, what we’re comparing the DataFrame to, can be a DataFrame, Series, dict, or array of values. See the docs for more.To get the rows where any of the conditions are met:
In [92]: dfi = DataFrame({'A': [1, 2, 3, 4], 'B': ['a', 'b', 'f', 'n']}) In [93]: dfi Out[93]: A B 0 1 a 1 2 b 2 3 f 3 4 n [4 rows x 2 columns] In [94]: other = DataFrame({'A': [1, 3, 3, 7], 'B': ['e', 'f', 'f', 'e']}) In [95]: mask = dfi.isin(other) In [96]: mask Out[96]: A B 0 True False 1 False False 2 True True 3 False False [4 rows x 2 columns] In [97]: dfi[mask.any(1)] ����������������������������������������������������������������������������������������������������������������Out[97]: A B 0 1 a 2 3 f [2 rows x 2 columns]
Seriesnow supports ato_framemethod to convert it to a single-column DataFrame (GH5164)All R datasets listed here http://stat.ethz.ch/R-manual/R-devel/library/datasets/html/00Index.html can now be loaded into Pandas objects
# note that pandas.rpy was deprecated in v0.16.0 import pandas.rpy.common as com com.load_data('Titanic')
tz_localizecan infer a fall daylight savings transition based on the structure of the unlocalized data (GH4230), see the docsDatetimeIndexis now in the API documentation, see the docsjson_normalize()is a new method to allow you to create a flat table from semi-structured JSON data. See the docs (GH1067)Added PySide support for the qtpandas DataFrameModel and DataFrameWidget.
Python csv parser now supports usecols (GH4335)
Frequencies gained several new offsets:
DataFrame has a new
interpolatemethod, similar to Series (GH4434, GH1892)In [98]: df = DataFrame({'A': [1, 2.1, np.nan, 4.7, 5.6, 6.8], ....: 'B': [.25, np.nan, np.nan, 4, 12.2, 14.4]}) ....: In [99]: df.interpolate() Out[99]: A B 0 1.0 0.25 1 2.1 1.50 2 3.4 2.75 3 4.7 4.00 4 5.6 12.20 5 6.8 14.40 [6 rows x 2 columns]
Additionally, the
methodargument tointerpolatehas been expanded to include'nearest', 'zero', 'slinear', 'quadratic', 'cubic', 'barycentric', 'krogh', 'piecewise_polynomial', 'pchip', `polynomial`, 'spline'The new methods require scipy. Consult the Scipy reference guide and documentation for more information about when the various methods are appropriate. See the docs.Interpolate now also accepts a
limitkeyword argument. This works similar tofillna‘s limit:In [100]: ser = Series([1, 3, np.nan, np.nan, np.nan, 11]) In [101]: ser.interpolate(limit=2) Out[101]: 0 1.0 1 3.0 2 5.0 3 7.0 4 NaN 5 11.0 Length: 6, dtype: float64
Added
wide_to_longpanel data convenience function. See the docs.In [102]: np.random.seed(123) In [103]: df = pd.DataFrame({"A1970" : {0 : "a", 1 : "b", 2 : "c"}, .....: "A1980" : {0 : "d", 1 : "e", 2 : "f"}, .....: "B1970" : {0 : 2.5, 1 : 1.2, 2 : .7}, .....: "B1980" : {0 : 3.2, 1 : 1.3, 2 : .1}, .....: "X" : dict(zip(range(3), np.random.randn(3))) .....: }) .....: In [104]: df["id"] = df.index In [105]: df Out[105]: A1970 A1980 B1970 B1980 X id 0 a d 2.5 3.2 -1.085631 0 1 b e 1.2 1.3 0.997345 1 2 c f 0.7 0.1 0.282978 2 [3 rows x 6 columns] In [106]: wide_to_long(df, ["A", "B"], i="id", j="year") ���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[106]: X A B id year 0 1970 -1.085631 a 2.5 1 1970 0.997345 b 1.2 2 1970 0.282978 c 0.7 0 1980 -1.085631 d 3.2 1 1980 0.997345 e 1.3 2 1980 0.282978 f 0.1 [6 rows x 3 columns]
to_csvnow takes adate_formatkeyword argument that specifies how output datetime objects should be formatted. Datetimes encountered in the index, columns, and values will all have this formatting applied. (GH4313)DataFrame.plotwill scatter plot x versus y by passingkind='scatter'(GH2215)- Added support for Google Analytics v3 API segment IDs that also supports v2 IDs. (GH5271)
Experimental¶
The new
eval()function implements expression evaluation usingnumexprbehind the scenes. This results in large speedups for complicated expressions involving large DataFrames/Series. For example,In [107]: nrows, ncols = 20000, 100 In [108]: df1, df2, df3, df4 = [DataFrame(randn(nrows, ncols)) .....: for _ in range(4)] .....:
# eval with NumExpr backend In [109]: %timeit pd.eval('df1 + df2 + df3 + df4') 7.17 ms +- 323 us per loop (mean +- std. dev. of 7 runs, 100 loops each)
# pure Python evaluation In [110]: %timeit df1 + df2 + df3 + df4 10.6 ms +- 598 us per loop (mean +- std. dev. of 7 runs, 100 loops each)
For more details, see the the docs
Similar to
pandas.eval,DataFramehas a newDataFrame.evalmethod that evaluates an expression in the context of theDataFrame. For example,In [111]: df = DataFrame(randn(10, 2), columns=['a', 'b']) In [112]: df.eval('a + b') Out[112]: 0 -0.685204 1 1.589745 2 0.325441 3 -1.784153 4 -0.432893 5 0.171850 6 1.895919 7 3.065587 8 -0.092759 9 1.391365 Length: 10, dtype: float64
query()method has been added that allows you to select elements of aDataFrameusing a natural query syntax nearly identical to Python syntax. For example,In [113]: n = 20 In [114]: df = DataFrame(np.random.randint(n, size=(n, 3)), columns=['a', 'b', 'c']) In [115]: df.query('a < b < c') Out[115]: a b c 11 1 5 8 15 8 16 19 [2 rows x 3 columns]
selects all the rows of
dfwherea < b < cevaluates toTrue. For more details see the the docs.pd.read_msgpack()andpd.to_msgpack()are now a supported method of serialization of arbitrary pandas (and python objects) in a lightweight portable binary format. See the docsWarning
Since this is an EXPERIMENTAL LIBRARY, the storage format may not be stable until a future release.
In [116]: df = DataFrame(np.random.rand(5,2),columns=list('AB')) In [117]: df.to_msgpack('foo.msg') In [118]: pd.read_msgpack('foo.msg') Out[118]: A B 0 0.251082 0.017357 1 0.347915 0.929879 2 0.546233 0.203368 3 0.064942 0.031722 4 0.355309 0.524575 [5 rows x 2 columns] In [119]: s = Series(np.random.rand(5),index=date_range('20130101',periods=5)) In [120]: pd.to_msgpack('foo.msg', df, s) In [121]: pd.read_msgpack('foo.msg') Out[121]: [ A B 0 0.251082 0.017357 1 0.347915 0.929879 2 0.546233 0.203368 3 0.064942 0.031722 4 0.355309 0.524575 [5 rows x 2 columns], 2013-01-01 0.022321 2013-01-02 0.227025 2013-01-03 0.383282 2013-01-04 0.193225 2013-01-05 0.110977 Freq: D, Length: 5, dtype: float64]
You can pass
iterator=Trueto iterator over the unpacked resultsIn [122]: for o in pd.read_msgpack('foo.msg',iterator=True): .....: print o .....: File "<ipython-input-122-59af9f4d3a62>", line 2 print o ^ SyntaxError: Missing parentheses in call to 'print'
pandas.io.gbqprovides a simple way to extract from, and load data into, Google’s BigQuery Data Sets by way of pandas DataFrames. BigQuery is a high performance SQL-like database service, useful for performing ad-hoc queries against extremely large datasets. See the docsfrom pandas.io import gbq # A query to select the average monthly temperatures in the # in the year 2000 across the USA. The dataset, # publicata:samples.gsod, is available on all BigQuery accounts, # and is based on NOAA gsod data. query = """SELECT station_number as STATION, month as MONTH, AVG(mean_temp) as MEAN_TEMP FROM publicdata:samples.gsod WHERE YEAR = 2000 GROUP BY STATION, MONTH ORDER BY STATION, MONTH ASC""" # Fetch the result set for this query # Your Google BigQuery Project ID # To find this, see your dashboard: # https://console.developers.google.com/iam-admin/projects?authuser=0 projectid = xxxxxxxxx; df = gbq.read_gbq(query, project_id = projectid) # Use pandas to process and reshape the dataset df2 = df.pivot(index='STATION', columns='MONTH', values='MEAN_TEMP') df3 = pandas.concat([df2.min(), df2.mean(), df2.max()], axis=1,keys=["Min Tem", "Mean Temp", "Max Temp"])
The resulting DataFrame is:
> df3 Min Tem Mean Temp Max Temp MONTH 1 -53.336667 39.827892 89.770968 2 -49.837500 43.685219 93.437932 3 -77.926087 48.708355 96.099998 4 -82.892858 55.070087 97.317240 5 -92.378261 61.428117 102.042856 6 -77.703334 65.858888 102.900000 7 -87.821428 68.169663 106.510714 8 -89.431999 68.614215 105.500000 9 -86.611112 63.436935 107.142856 10 -78.209677 56.880838 92.103333 11 -50.125000 48.861228 94.996428 12 -50.332258 42.286879 94.396774
Warning
To use this module, you will need a BigQuery account. See <https://cloud.google.com/products/big-query> for details.
As of 10/10/13, there is a bug in Google’s API preventing result sets from being larger than 100,000 rows. A patch is scheduled for the week of 10/14/13.
Internal Refactoring¶
In 0.13.0 there is a major refactor primarily to subclass Series from
NDFrame, which is the base class currently for DataFrame and Panel,
to unify methods and behaviors. Series formerly subclassed directly from
ndarray. (GH4080, GH3862, GH816)
Warning
There are two potential incompatibilities from < 0.13.0
Using certain numpy functions would previously return a
Seriesif passed aSeriesas an argument. This seems only to affectnp.ones_like,np.empty_like,np.diffandnp.where. These now returnndarrays.In [123]: s = Series([1,2,3,4])
Numpy Usage
In [124]: np.ones_like(s) Out[124]: array([1, 1, 1, 1]) In [125]: np.diff(s) ������������������������������Out[125]: array([1, 1, 1]) In [126]: np.where(s>1,s,np.nan) ���������������������������������������������������������Out[126]: array([ nan, 2., 3., 4.])
Pandonic Usage
In [127]: Series(1,index=s.index) Out[127]: 0 1 1 1 2 1 3 1 Length: 4, dtype: int64 In [128]: s.diff() ���������������������������������������������������������������Out[128]: 0 NaN 1 1.0 2 1.0 3 1.0 Length: 4, dtype: float64 In [129]: s.where(s>1) ����������������������������������������������������������������������������������������������������������������������������������������Out[129]: 0 NaN 1 2.0 2 3.0 3 4.0 Length: 4, dtype: float64
Passing a
Seriesdirectly to a cython function expecting anndarraytype will no long work directly, you must passSeries.values, See Enhancing PerformanceSeries(0.5)would previously return the scalar0.5, instead this will return a 1-elementSeriesThis change breaks
rpy2<=2.3.8. an Issue has been opened against rpy2 and a workaround is detailed in GH5698. Thanks @JanSchulz.
Pickle compatibility is preserved for pickles created prior to 0.13. These must be unpickled with
pd.read_pickle, see Pickling.Refactor of series.py/frame.py/panel.py to move common code to generic.py
- added
_setup_axesto created generic NDFrame structures - moved methods
from_axes,_wrap_array,axes,ix,loc,iloc,shape,empty,swapaxes,transpose,pop__iter__,keys,__contains__,__len__,__neg__,__invert__convert_objects,as_blocks,as_matrix,values__getstate__,__setstate__(compat remains in frame/panel)__getattr__,__setattr___indexed_same,reindex_like,align,where,maskfillna,replace(Seriesreplace is now consistent withDataFrame)filter(also added axis argument to selectively filter on a different axis)reindex,reindex_axis,taketruncate(moved to become part ofNDFrame)
- added
These are API changes which make
Panelmore consistent withDataFrameswapaxeson aPanelwith the same axes specified now return a copy- support attribute access for setting
- filter supports the same API as the original
DataFramefilter
Reindex called with no arguments will now return a copy of the input object
TimeSeriesis now an alias forSeries. the propertyis_time_seriescan be used to distinguish (if desired)Refactor of Sparse objects to use BlockManager
- Created a new block type in internals,
SparseBlock, which can hold multi-dtypes and is non-consolidatable.SparseSeriesandSparseDataFramenow inherit more methods from there hierarchy (Series/DataFrame), and no longer inherit fromSparseArray(which instead is the object of theSparseBlock) - Sparse suite now supports integration with non-sparse data. Non-float sparse data is supportable (partially implemented)
- Operations on sparse structures within DataFrames should preserve sparseness, merging type operations will convert to dense (and back to sparse), so might be somewhat inefficient
- enable setitem on
SparseSeriesfor boolean/integer/slices SparsePanelsimplementation is unchanged (e.g. not using BlockManager, needs work)
- Created a new block type in internals,
added
ftypesmethod to Series/DataFrame, similar todtypes, but indicates if the underlying is sparse/dense (as well as the dtype)All
NDFrameobjects can now use__finalize__()to specify various values to propagate to new objects from an existing one (e.g.nameinSerieswill follow more automatically now)Internal type checking is now done via a suite of generated classes, allowing
isinstance(value, klass)without having to directly import the klass, courtesy of @jtratnerBug in Series update where the parent frame is not updating its cache based on changes (GH4080) or types (GH3217), fillna (GH3386)
Refactor
Series.reindexto core/generic.py (GH4604, GH4618), allowmethod=in reindexing on a Series to workSeries.copyno longer accepts theorderparameter and is now consistent withNDFramecopyRefactor
renamemethods to core/generic.py; fixesSeries.renamefor (GH4605), and addsrenamewith the same signature forPanelRefactor
clipmethods to core/generic.py (GH4798)Refactor of
_get_numeric_data/_get_bool_datato core/generic.py, allowing Series/Panel functionalitySeries(for index) /Panel(for items) now allow attribute access to its elements (GH1903)In [130]: s = Series([1,2,3],index=list('abc')) In [131]: s.b Out[131]: 2 In [132]: s.a = 5 In [133]: s Out[133]: a 5 b 2 c 3 Length: 3, dtype: int64
Bug Fixes¶
See V0.13.0 Bug Fixes for an extensive list of bugs that have been fixed in 0.13.0.
See the full release notes or issue tracker on GitHub for a complete list of all API changes, Enhancements and Bug Fixes.
v0.12.0 (July 24, 2013)¶
This is a major release from 0.11.0 and includes several new features and enhancements along with a large number of bug fixes.
Highlights include a consistent I/O API naming scheme, routines to read html,
write multi-indexes to csv files, read & write STATA data files, read & write JSON format
files, Python 3 support for HDFStore, filtering of groupby expressions via filter, and a
revamped replace routine that accepts regular expressions.
API changes¶
The I/O API is now much more consistent with a set of top level
readerfunctions accessed likepd.read_csv()that generally return apandasobject.
read_csvread_excelread_hdfread_sqlread_jsonread_htmlread_stataread_clipboardThe corresponding
writerfunctions are object methods that are accessed likedf.to_csv()
to_csvto_excelto_hdfto_sqlto_jsonto_htmlto_statato_clipboardFix modulo and integer division on Series,DataFrames to act similary to
floatdtypes to returnnp.nanornp.infas appropriate (GH3590). This correct a numpy bug that treatsintegerandfloatdtypes differently.In [1]: p = DataFrame({ 'first' : [4,5,8], 'second' : [0,0,3] }) In [2]: p % 0 Out[2]: first second 0 NaN NaN 1 NaN NaN 2 NaN NaN [3 rows x 2 columns] In [3]: p % p ���������������������������������������������������������������������������������������������������Out[3]: first second 0 0.0 NaN 1 0.0 NaN 2 0.0 0.0 [3 rows x 2 columns] In [4]: p / p ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[4]: first second 0 1.0 NaN 1 1.0 NaN 2 1.0 1.0 [3 rows x 2 columns] In [5]: p / 0 ���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[5]: first second 0 inf NaN 1 inf NaN 2 inf inf [3 rows x 2 columns]Add
squeezekeyword togroupbyto allow reduction from DataFrame -> Series if groups are unique. This is a Regression from 0.10.1. We are reverting back to the prior behavior. This means groupby will return the same shaped objects whether the groups are unique or not. Revert this issue (GH2893) with (GH3596).In [6]: df2 = DataFrame([{"val1": 1, "val2" : 20}, {"val1":1, "val2": 19}, ...: {"val1":1, "val2": 27}, {"val1":1, "val2": 12}]) ...: In [7]: def func(dataf): ...: return dataf["val2"] - dataf["val2"].mean() ...: # squeezing the result frame to a series (because we have unique groups) In [8]: df2.groupby("val1", squeeze=True).apply(func) Out[8]: 0 0.5 1 -0.5 2 7.5 3 -7.5 Name: 1, Length: 4, dtype: float64 # no squeezing (the default, and behavior in 0.10.1) In [9]: df2.groupby("val1").apply(func) ��������������������������������������������������������������������������������Out[9]: val2 0 1 2 3 val1 1 0.5 -0.5 7.5 -7.5 [1 rows x 4 columns]Raise on
ilocwhen boolean indexing with a label based indexer mask e.g. a boolean Series, even with integer labels, will raise. Sinceilocis purely positional based, the labels on the Series are not alignable (GH3631)This case is rarely used, and there are plently of alternatives. This preserves the
ilocAPI to be purely positional based.In [10]: df = DataFrame(lrange(5), list('ABCDE'), columns=['a']) In [11]: mask = (df.a%2 == 0) In [12]: mask Out[12]: A True B False C True D False E True Name: a, Length: 5, dtype: bool # this is what you should use In [13]: df.loc[mask] �������������������������������������������������������������������������������������������������Out[13]: a A 0 C 2 E 4 [3 rows x 1 columns] # this will work as well In [14]: df.iloc[mask.values] �����������������������������������������������������������������������������������������������������������������������������������������������������Out[14]: a A 0 C 2 E 4 [3 rows x 1 columns]
df.iloc[mask]will raise aValueErrorThe
raise_on_errorargument to plotting functions is removed. Instead, plotting functions raise aTypeErrorwhen thedtypeof the object isobjectto remind you to avoidobjectarrays whenever possible and thus you should cast to an appropriate numeric dtype if you need to plot something.Add
colormapkeyword to DataFrame plotting methods. Accepts either a matplotlib colormap object (ie, matplotlib.cm.jet) or a string name of such an object (ie, ‘jet’). The colormap is sampled to select the color for each column. Please see Colormaps for more information. (GH3860)
DataFrame.interpolate()is now deprecated. Please useDataFrame.fillna()andDataFrame.replace()instead. (GH3582, GH3675, GH3676)the
methodandaxisarguments ofDataFrame.replace()are deprecated
DataFrame.replace‘sinfer_typesparameter is removed and now performs conversion by default. (GH3907)Add the keyword
allow_duplicatestoDataFrame.insertto allow a duplicate column to be inserted ifTrue, default isFalse(same as prior to 0.12) (GH3679)IO api
added top-level function
read_excelto replace the following, The original API is deprecated and will be removed in a future versionfrom pandas.io.parsers import ExcelFile xls = ExcelFile('path_to_file.xls') xls.parse('Sheet1', index_col=None, na_values=['NA'])With
import pandas as pd pd.read_excel('path_to_file.xls', 'Sheet1', index_col=None, na_values=['NA'])added top-level function
read_sqlthat is equivalent to the followingfrom pandas.io.sql import read_frame read_frame(....)
DataFrame.to_htmlandDataFrame.to_latexnow accept a path for their first argument (GH3702)Do not allow astypes on
datetime64[ns]except toobject, andtimedelta64[ns]toobject/int(GH3425)The behavior of
datetime64dtypes has changed with respect to certain so-called reduction operations (GH3726). The following operations now raise aTypeErrorwhen perfomed on aSeriesand return an emptySerieswhen performed on aDataFramesimilar to performing these operations on, for example, aDataFrameofsliceobjects:
- sum, prod, mean, std, var, skew, kurt, corr, and cov
read_htmlnow defaults toNonewhen reading, and falls back onbs4+html5libwhen lxml fails to parse. a list of parsers to try until success is also validThe internal
pandasclass hierarchy has changed (slightly). The previousPandasObjectnow is calledPandasContainerand a newPandasObjecthas become the baseclass forPandasContaineras well asIndex,Categorical,GroupBy,SparseList, andSparseArray(+ their base classes). Currently,PandasObjectprovides string methods (fromStringMixin). (GH4090, GH4092)New
StringMixinthat, given a__unicode__method, gets python 2 and python 3 compatible string methods (__str__,__bytes__, and__repr__). Plus string safety throughout. Now employed in many places throughout the pandas library. (GH4090, GH4092)
I/O Enhancements¶
pd.read_html()can now parse HTML strings, files or urls and return DataFrames, courtesy of @cpcloud. (GH3477, GH3605, GH3606, GH3616). It works with a single parser backend: BeautifulSoup4 + html5lib See the docsYou can use
pd.read_html()to read the output fromDataFrame.to_html()like soIn [15]: df = DataFrame({'a': range(3), 'b': list('abc')}) In [16]: print(df) a b 0 0 a 1 1 b 2 2 c [3 rows x 2 columns] In [17]: html = df.to_html() In [18]: alist = pd.read_html(html, index_col=0) In [19]: print(df == alist[0]) a b 0 True True 1 True True 2 True True [3 rows x 2 columns]Note that
alisthere is a Pythonlistsopd.read_html()andDataFrame.to_html()are not inverses.
pd.read_html()no longer performs hard conversion of date strings (GH3656).Warning
You may have to install an older version of BeautifulSoup4, See the installation docs
Added module for reading and writing Stata files:
pandas.io.stata(GH1512) accessable viaread_statatop-level function for reading, andto_stataDataFrame method for writing, See the docsAdded module for reading and writing json format files:
pandas.io.jsonaccessable viaread_jsontop-level function for reading, andto_jsonDataFrame method for writing, See the docs various issues (GH1226, GH3804, GH3876, GH3867, GH1305)
MultiIndexcolumn support for reading and writing csv format files
The
headeroption inread_csvnow accepts a list of the rows from which to read the index.The option,
tupleize_colscan now be specified in bothto_csvandread_csv, to provide compatiblity for the pre 0.12 behavior of writing and readingMultIndexcolumns via a list of tuples. The default in 0.12 is to write lists of tuples and not interpret list of tuples as aMultiIndexcolumn.Note: The default behavior in 0.12 remains unchanged from prior versions, but starting with 0.13, the default to write and read
MultiIndexcolumns will be in the new format. (GH3571, GH1651, GH3141)If an
index_colis not specified (e.g. you don’t have an index, or wrote it withdf.to_csv(..., index=False), then anynameson the columns index will be lost.In [20]: from pandas.util.testing import makeCustomDataframe as mkdf In [21]: df = mkdf(5,3,r_idx_nlevels=2,c_idx_nlevels=4) In [22]: df.to_csv('mi.csv',tupleize_cols=False) In [23]: print(open('mi.csv').read()) C0,,C_l0_g0,C_l0_g1,C_l0_g2 C1,,C_l1_g0,C_l1_g1,C_l1_g2 C2,,C_l2_g0,C_l2_g1,C_l2_g2 C3,,C_l3_g0,C_l3_g1,C_l3_g2 R0,R1,,, R_l0_g0,R_l1_g0,R0C0,R0C1,R0C2 R_l0_g1,R_l1_g1,R1C0,R1C1,R1C2 R_l0_g2,R_l1_g2,R2C0,R2C1,R2C2 R_l0_g3,R_l1_g3,R3C0,R3C1,R3C2 R_l0_g4,R_l1_g4,R4C0,R4C1,R4C2 In [24]: pd.read_csv('mi.csv',header=[0,1,2,3],index_col=[0,1],tupleize_cols=False) �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[24]: C0 C_l0_g0 C_l0_g1 C_l0_g2 C1 C_l1_g0 C_l1_g1 C_l1_g2 C2 C_l2_g0 C_l2_g1 C_l2_g2 C3 C_l3_g0 C_l3_g1 C_l3_g2 R0 R1 R_l0_g0 R_l1_g0 R0C0 R0C1 R0C2 R_l0_g1 R_l1_g1 R1C0 R1C1 R1C2 R_l0_g2 R_l1_g2 R2C0 R2C1 R2C2 R_l0_g3 R_l1_g3 R3C0 R3C1 R3C2 R_l0_g4 R_l1_g4 R4C0 R4C1 R4C2 [5 rows x 3 columns]Support for
HDFStore(viaPyTables 3.0.0) on Python3Iterator support via
read_hdfthat automatically opens and closes the store when iteration is finished. This is only for tablesIn [25]: path = 'store_iterator.h5' In [26]: DataFrame(randn(10,2)).to_hdf(path,'df',table=True) In [27]: for df in read_hdf(path,'df', chunksize=3): ....: print df ....: 0 1 0 0.713216 -0.778461 1 -0.661062 0.862877 2 0.344342 0.149565 0 1 3 -0.626968 -0.875772 4 -0.930687 -0.218983 5 0.949965 -0.442354 0 1 6 -0.402985 1.111358 7 -0.241527 -0.670477 8 0.049355 0.632633 0 1 9 -1.502767 -1.225492
read_csvwill now throw a more informative error message when a file contains no columns, e.g., all newline characters
Other Enhancements¶
DataFrame.replace()now allows regular expressions on containedSerieswith object dtype. See the examples section in the regular docs Replacing via String ExpressionFor example you can do
In [25]: df = DataFrame({'a': list('ab..'), 'b': [1, 2, 3, 4]}) In [26]: df.replace(regex=r'\s*\.\s*', value=np.nan) Out[26]: a b 0 a 1 1 b 2 2 NaN 3 3 NaN 4 [4 rows x 2 columns]to replace all occurrences of the string
'.'with zero or more instances of surrounding whitespace withNaN.Regular string replacement still works as expected. For example, you can do
In [27]: df.replace('.', np.nan) Out[27]: a b 0 a 1 1 b 2 2 NaN 3 3 NaN 4 [4 rows x 2 columns]to replace all occurrences of the string
'.'withNaN.
pd.melt()now accepts the optional parametersvar_nameandvalue_nameto specify custom column names of the returned DataFrame.
pd.set_option()now allows N option, value pairs (GH3667).Let’s say that we had an option
'a.b'and another option'b.c'. We can set them at the same time:In [28]: pd.get_option('a.b') Out[28]: 2 In [29]: pd.get_option('b.c') �����������Out[29]: 3 In [30]: pd.set_option('a.b', 1, 'b.c', 4) In [31]: pd.get_option('a.b') Out[31]: 1 In [32]: pd.get_option('b.c') �����������Out[32]: 4The
filtermethod for group objects returns a subset of the original object. Suppose we want to take only elements that belong to groups with a group sum greater than 2.In [33]: sf = Series([1, 1, 2, 3, 3, 3]) In [34]: sf.groupby(sf).filter(lambda x: x.sum() > 2) Out[34]: 3 3 4 3 5 3 Length: 3, dtype: int64The argument of
filtermust a function that, applied to the group as a whole, returnsTrueorFalse.Another useful operation is filtering out elements that belong to groups with only a couple members.
In [35]: dff = DataFrame({'A': np.arange(8), 'B': list('aabbbbcc')}) In [36]: dff.groupby('B').filter(lambda x: len(x) > 2) Out[36]: A B 2 2 b 3 3 b 4 4 b 5 5 b [4 rows x 2 columns]Alternatively, instead of dropping the offending groups, we can return a like-indexed objects where the groups that do not pass the filter are filled with NaNs.
In [37]: dff.groupby('B').filter(lambda x: len(x) > 2, dropna=False) Out[37]: A B 0 NaN NaN 1 NaN NaN 2 2.0 b 3 3.0 b 4 4.0 b 5 5.0 b 6 NaN NaN 7 NaN NaN [8 rows x 2 columns]Series and DataFrame hist methods now take a
figsizeargument (GH3834)DatetimeIndexes no longer try to convert mixed-integer indexes during join operations (GH3877)
Timestamp.min and Timestamp.max now represent valid Timestamp instances instead of the default datetime.min and datetime.max (respectively), thanks @SleepingPills
read_htmlnow raises when no tables are found and BeautifulSoup==4.2.0 is detected (GH4214)
Experimental Features¶
Added experimental
CustomBusinessDayclass to supportDateOffsetswith custom holiday calendars and custom weekmasks. (GH2301)Note
This uses the
numpy.busdaycalendarAPI introduced in Numpy 1.7 and therefore requires Numpy 1.7.0 or newer.In [38]: from pandas.tseries.offsets import CustomBusinessDay In [39]: from datetime import datetime # As an interesting example, let's look at Egypt where # a Friday-Saturday weekend is observed. In [40]: weekmask_egypt = 'Sun Mon Tue Wed Thu' # They also observe International Workers' Day so let's # add that for a couple of years In [41]: holidays = ['2012-05-01', datetime(2013, 5, 1), np.datetime64('2014-05-01')] In [42]: bday_egypt = CustomBusinessDay(holidays=holidays, weekmask=weekmask_egypt) In [43]: dt = datetime(2013, 4, 30) In [44]: print(dt + 2 * bday_egypt) 2013-05-05 00:00:00 In [45]: dts = date_range(dt, periods=5, freq=bday_egypt) In [46]: print(Series(dts.weekday, dts).map(Series('Mon Tue Wed Thu Fri Sat Sun'.split()))) 2013-04-30 Tue 2013-05-02 Thu 2013-05-05 Sun 2013-05-06 Mon 2013-05-07 Tue Freq: C, Length: 5, dtype: object
Bug Fixes¶
Plotting functions now raise a
TypeErrorbefore trying to plot anything if the associated objects have have a dtype ofobject(GH1818, GH3572, GH3911, GH3912), but they will try to convert object arrays to numeric arrays if possible so that you can still plot, for example, an object array with floats. This happens before any drawing takes place which elimnates any spurious plots from showing up.
fillnamethods now raise aTypeErrorif thevalueparameter is a list or tuple.
Series.strnow supports iteration (GH3638). You can iterate over the individual elements of each string in theSeries. Each iteration yields yields aSerieswith either a single character at each index of the originalSeriesorNaN. For example,In [47]: strs = 'go', 'bow', 'joe', 'slow' In [48]: ds = Series(strs) In [49]: for s in ds.str: ....: print(s) ....: 0 g 1 b 2 j 3 s Length: 4, dtype: object 0 o 1 o 2 o 3 l Length: 4, dtype: object 0 NaN 1 w 2 e 3 o Length: 4, dtype: object 0 NaN 1 NaN 2 NaN 3 w Length: 4, dtype: object In [50]: s ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[50]: 0 NaN 1 NaN 2 NaN 3 w Length: 4, dtype: object In [51]: s.dropna().values.item() == 'w' �����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[51]: TrueThe last element yielded by the iterator will be a
Seriescontaining the last element of the longest string in theSerieswith all other elements beingNaN. Here since'slow'is the longest string and there are no other strings with the same length'w'is the only non-null string in the yieldedSeries.
HDFStore
- will retain index attributes (freq,tz,name) on recreation (GH3499)
- will warn with a
AttributeConflictWarningif you are attempting to append an index with a different frequency than the existing, or attempting to append an index with a different name than the existing- support datelike columns with a timezone as data_columns (GH2852)
Non-unique index support clarified (GH3468).
- Fix assigning a new index to a duplicate index in a DataFrame would fail (GH3468)
- Fix construction of a DataFrame with a duplicate index
- ref_locs support to allow duplicative indices across dtypes, allows iget support to always find the index (even across dtypes) (GH2194)
- applymap on a DataFrame with a non-unique index now works (removed warning) (GH2786), and fix (GH3230)
- Fix to_csv to handle non-unique columns (GH3495)
- Duplicate indexes with getitem will return items in the correct order (GH3455, GH3457) and handle missing elements like unique indices (GH3561)
- Duplicate indexes with and empty DataFrame.from_records will return a correct frame (GH3562)
- Concat to produce a non-unique columns when duplicates are across dtypes is fixed (GH3602)
- Allow insert/delete to non-unique columns (GH3679)
- Non-unique indexing with a slice via
locand friends fixed (GH3659)- Allow insert/delete to non-unique columns (GH3679)
- Extend
reindexto correctly deal with non-unique indices (GH3679)DataFrame.itertuples()now works with frames with duplicate column names (GH3873)- Bug in non-unique indexing via
iloc(GH4017); addedtakeableargument toreindexfor location-based taking- Allow non-unique indexing in series via
.ix/.locand__getitem__(GH4246)- Fixed non-unique indexing memory allocation issue with
.ix/.loc(GH4280)
DataFrame.from_recordsdid not accept empty recarrays (GH3682)
read_htmlnow correctly skips tests (GH3741)Fixed a bug where
DataFrame.replacewith a compiled regular expression in theto_replaceargument wasn’t working (GH3907)Improved
networktest decorator to catchIOError(and thereforeURLErroras well). Addedwith_connectivity_checkdecorator to allow explicitly checking a website as a proxy for seeing if there is network connectivity. Plus, newoptional_argsdecorator factory for decorators. (GH3910, GH3914)Fixed testing issue where too many sockets where open thus leading to a connection reset issue (GH3982, GH3985, GH4028, GH4054)
Fixed failing tests in test_yahoo, test_google where symbols were not retrieved but were being accessed (GH3982, GH3985, GH4028, GH4054)
Series.histwill now take the figure from the current environment if one is not passedFixed bug where a 1xN DataFrame would barf on a 1xN mask (GH4071)
Fixed running of
toxunder python3 where the pickle import was getting rewritten in an incompatible way (GH4062, GH4063)Fixed bug where sharex and sharey were not being passed to grouped_hist (GH4089)
Fixed bug in
DataFrame.replacewhere a nested dict wasn’t being iterated over when regex=False (GH4115)Fixed bug in the parsing of microseconds when using the
formatargument into_datetime(GH4152)Fixed bug in
PandasAutoDateLocatorwhereinvert_xaxistriggered incorrectlyMilliSecondLocator(GH3990)Fixed bug in plotting that wasn’t raising on invalid colormap for matplotlib 1.1.1 (GH4215)
Fixed the legend displaying in
DataFrame.plot(kind='kde')(GH4216)Fixed bug where Index slices weren’t carrying the name attribute (GH4226)
Fixed bug in initializing
DatetimeIndexwith an array of strings in a certain time zone (GH4229)Fixed bug where html5lib wasn’t being properly skipped (GH4265)
Fixed bug where get_data_famafrench wasn’t using the correct file edges (GH4281)
See the full release notes or issue tracker on GitHub for a complete list.
v0.11.0 (April 22, 2013)¶
This is a major release from 0.10.1 and includes many new features and enhancements along with a large number of bug fixes. The methods of Selecting Data have had quite a number of additions, and Dtype support is now full-fledged. There are also a number of important API changes that long-time pandas users should pay close attention to.
There is a new section in the documentation, 10 Minutes to Pandas, primarily geared to new users.
There is a new section in the documentation, Cookbook, a collection of useful recipes in pandas (and that we want contributions!).
There are several libraries that are now Recommended Dependencies
Selection Choices¶
Starting in 0.11.0, object selection has had a number of user-requested additions in order to support more explicit location based indexing. Pandas now supports three types of multi-axis indexing.
.locis strictly label based, will raiseKeyErrorwhen the items are not found, allowed inputs are:- A single label, e.g.
5or'a', (note that5is interpreted as a label of the index. This use is not an integer position along the index) - A list or array of labels
['a', 'b', 'c'] - A slice object with labels
'a':'f', (note that contrary to usual python slices, both the start and the stop are included!) - A boolean array
See more at Selection by Label
- A single label, e.g.
.ilocis strictly integer position based (from0tolength-1of the axis), will raiseIndexErrorwhen the requested indicies are out of bounds. Allowed inputs are:- An integer e.g.
5 - A list or array of integers
[4, 3, 0] - A slice object with ints
1:7 - A boolean array
See more at Selection by Position
- An integer e.g.
.ixsupports mixed integer and label based access. It is primarily label based, but will fallback to integer positional access..ixis the most general and will support any of the inputs to.locand.iloc, as well as support for floating point label schemes..ixis especially useful when dealing with mixed positional and label based hierarchial indexes.As using integer slices with
.ixhave different behavior depending on whether the slice is interpreted as position based or label based, it’s usually better to be explicit and use.ilocor.loc.See more at Advanced Indexing and Advanced Hierarchical.
Selection Deprecations¶
Starting in version 0.11.0, these methods may be deprecated in future versions.
irowicoliget_value
See the section Selection by Position for substitutes.
Dtypes¶
Numeric dtypes will propagate and can coexist in DataFrames. If a dtype is passed (either directly via the dtype keyword, a passed ndarray, or a passed Series, then it will be preserved in DataFrame operations. Furthermore, different numeric dtypes will NOT be combined. The following example will give you a taste.
In [1]: df1 = DataFrame(randn(8, 1), columns = ['A'], dtype = 'float32')
In [2]: df1
Out[2]:
A
0 1.392665
1 -0.123497
2 -0.402761
3 -0.246604
4 -0.288433
5 -0.763434
6 2.069526
7 -1.203569
[8 rows x 1 columns]
In [3]: df1.dtypes
�������������������������������������������������������������������������������������������������������������������������������������������Out[3]:
A float32
Length: 1, dtype: object
In [4]: df2 = DataFrame(dict( A = Series(randn(8),dtype='float16'),
...: B = Series(randn(8)),
...: C = Series(randn(8),dtype='uint8') ))
...:
In [5]: df2
Out[5]:
A B C
0 0.591797 -0.038605 0
1 0.841309 -0.460478 1
2 -0.500977 -0.310458 0
3 -0.816406 0.866493 254
4 -0.207031 0.245972 0
5 -0.664062 0.319442 1
6 0.580566 1.378512 1
7 -0.965820 0.292502 255
[8 rows x 3 columns]
In [6]: df2.dtypes
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[6]:
A float16
B float64
C uint8
Length: 3, dtype: object
# here you get some upcasting
In [7]: df3 = df1.reindex_like(df2).fillna(value=0.0) + df2
In [8]: df3
Out[8]:
A B C
0 1.984462 -0.038605 0.0
1 0.717812 -0.460478 1.0
2 -0.903737 -0.310458 0.0
3 -1.063011 0.866493 254.0
4 -0.495465 0.245972 0.0
5 -1.427497 0.319442 1.0
6 2.650092 1.378512 1.0
7 -2.169390 0.292502 255.0
[8 rows x 3 columns]
In [9]: df3.dtypes
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[9]:
A float32
B float64
C float64
Length: 3, dtype: object
Dtype Conversion¶
This is lower-common-denomicator upcasting, meaning you get the dtype which can accomodate all of the types
In [10]: df3.values.dtype
Out[10]: dtype('float64')
Conversion
In [11]: df3.astype('float32').dtypes
Out[11]:
A float32
B float32
C float32
Length: 3, dtype: object
Mixed Conversion
In [12]: df3['D'] = '1.'
In [13]: df3['E'] = '1'
In [14]: df3.convert_objects(convert_numeric=True).dtypes
Out[14]:
A float32
B float64
C float64
D float64
E int64
Length: 5, dtype: object
# same, but specific dtype conversion
In [15]: df3['D'] = df3['D'].astype('float16')
In [16]: df3['E'] = df3['E'].astype('int32')
In [17]: df3.dtypes
Out[17]:
A float32
B float64
C float64
D float16
E int32
Length: 5, dtype: object
Forcing Date coercion (and setting NaT when not datelike)
In [18]: from datetime import datetime
In [19]: s = Series([datetime(2001,1,1,0,0), 'foo', 1.0, 1,
....: Timestamp('20010104'), '20010105'],dtype='O')
....:
In [20]: s.convert_objects(convert_dates='coerce')
Out[20]:
0 2001-01-01
1 NaT
2 NaT
3 NaT
4 2001-01-04
5 2001-01-05
Length: 6, dtype: datetime64[ns]
Dtype Gotchas¶
Platform Gotchas
Starting in 0.11.0, construction of DataFrame/Series will use default dtypes of int64 and float64,
regardless of platform. This is not an apparent change from earlier versions of pandas. If you specify
dtypes, they WILL be respected, however (GH2837)
The following will all result in int64 dtypes
In [21]: DataFrame([1,2],columns=['a']).dtypes
Out[21]:
a int64
Length: 1, dtype: object
In [22]: DataFrame({'a' : [1,2] }).dtypes
����������������������������������������������Out[22]:
a int64
Length: 1, dtype: object
In [23]: DataFrame({'a' : 1 }, index=range(2)).dtypes
��������������������������������������������������������������������������������������������Out[23]:
a int64
Length: 1, dtype: object
Keep in mind that DataFrame(np.array([1,2])) WILL result in int32 on 32-bit platforms!
Upcasting Gotchas
Performing indexing operations on integer type data can easily upcast the data.
The dtype of the input data will be preserved in cases where nans are not introduced.
In [24]: dfi = df3.astype('int32')
In [25]: dfi['D'] = dfi['D'].astype('int64')
In [26]: dfi
Out[26]:
A B C D E
0 1 0 0 1 1
1 0 0 1 1 1
2 0 0 0 1 1
3 -1 0 254 1 1
4 0 0 0 1 1
5 -1 0 1 1 1
6 2 1 1 1 1
7 -2 0 255 1 1
[8 rows x 5 columns]
In [27]: dfi.dtypes
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[27]:
A int32
B int32
C int32
D int64
E int32
Length: 5, dtype: object
In [28]: casted = dfi[dfi>0]
In [29]: casted
Out[29]:
A B C D E
0 1.0 NaN NaN 1 1
1 NaN NaN 1.0 1 1
2 NaN NaN NaN 1 1
3 NaN NaN 254.0 1 1
4 NaN NaN NaN 1 1
5 NaN NaN 1.0 1 1
6 2.0 1.0 1.0 1 1
7 NaN NaN 255.0 1 1
[8 rows x 5 columns]
In [30]: casted.dtypes
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[30]:
A float64
B float64
C float64
D int64
E int32
Length: 5, dtype: object
While float dtypes are unchanged.
In [31]: df4 = df3.copy()
In [32]: df4['A'] = df4['A'].astype('float32')
In [33]: df4.dtypes
Out[33]:
A float32
B float64
C float64
D float16
E int32
Length: 5, dtype: object
In [34]: casted = df4[df4>0]
In [35]: casted
Out[35]:
A B C D E
0 1.984462 NaN NaN 1.0 1
1 0.717812 NaN 1.0 1.0 1
2 NaN NaN NaN 1.0 1
3 NaN 0.866493 254.0 1.0 1
4 NaN 0.245972 NaN 1.0 1
5 NaN 0.319442 1.0 1.0 1
6 2.650092 1.378512 1.0 1.0 1
7 NaN 0.292502 255.0 1.0 1
[8 rows x 5 columns]
In [36]: casted.dtypes
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[36]:
A float32
B float64
C float64
D float16
E int32
Length: 5, dtype: object
Datetimes Conversion¶
Datetime64[ns] columns in a DataFrame (or a Series) allow the use of np.nan to indicate a nan value,
in addition to the traditional NaT, or not-a-time. This allows convenient nan setting in a generic way.
Furthermore datetime64[ns] columns are created by default, when passed datetimelike objects (this change was introduced in 0.10.1)
(GH2809, GH2810)
In [37]: df = DataFrame(randn(6,2),date_range('20010102',periods=6),columns=['A','B'])
In [38]: df['timestamp'] = Timestamp('20010103')
In [39]: df
Out[39]:
A B timestamp
2001-01-02 1.023958 0.660103 2001-01-03
2001-01-03 1.236475 -2.170629 2001-01-03
2001-01-04 -0.270630 -1.685677 2001-01-03
2001-01-05 -0.440747 -0.115070 2001-01-03
2001-01-06 -0.632102 -0.585977 2001-01-03
2001-01-07 -1.444787 -0.201135 2001-01-03
[6 rows x 3 columns]
# datetime64[ns] out of the box
In [40]: df.get_dtype_counts()
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[40]:
datetime64[ns] 1
float64 2
Length: 2, dtype: int64
# use the traditional nan, which is mapped to NaT internally
In [41]: df.loc[df.index[2:4], ['A','timestamp']] = np.nan
In [42]: df
Out[42]:
A B timestamp
2001-01-02 1.023958 0.660103 2001-01-03
2001-01-03 1.236475 -2.170629 2001-01-03
2001-01-04 NaN -1.685677 NaT
2001-01-05 NaN -0.115070 NaT
2001-01-06 -0.632102 -0.585977 2001-01-03
2001-01-07 -1.444787 -0.201135 2001-01-03
[6 rows x 3 columns]
Astype conversion on datetime64[ns] to object, implicity converts NaT to np.nan
In [43]: import datetime
In [44]: s = Series([datetime.datetime(2001, 1, 2, 0, 0) for i in range(3)])
In [45]: s.dtype
Out[45]: dtype('<M8[ns]')
In [46]: s[1] = np.nan
In [47]: s
Out[47]:
0 2001-01-02
1 NaT
2 2001-01-02
Length: 3, dtype: datetime64[ns]
In [48]: s.dtype
����������������������������������������������������������������������������������������Out[48]: dtype('<M8[ns]')
In [49]: s = s.astype('O')
In [50]: s
Out[50]:
0 2001-01-02 00:00:00
1 NaT
2 2001-01-02 00:00:00
Length: 3, dtype: object
In [51]: s.dtype
��������������������������������������������������������������������������������������������������������������Out[51]: dtype('O')
API changes¶
- Added to_series() method to indicies, to facilitate the creation of indexers (GH3275)
HDFStore
- added the method
select_columnto select a single column from a table as a Series.- deprecated the
uniquemethod, can be replicated byselect_column(key,column).unique()min_itemsizeparameter toappendwill now automatically create data_columns for passed keys
Enhancements¶
Improved performance of df.to_csv() by up to 10x in some cases. (GH3059)
Numexpr is now a Recommended Dependencies, to accelerate certain types of numerical and boolean operations
Bottleneck is now a Recommended Dependencies, to accelerate certain types of
nanoperations
HDFStore
support
read_hdf/to_hdfAPI similar toread_csv/to_csvIn [52]: df = DataFrame(dict(A=lrange(5), B=lrange(5))) In [53]: df.to_hdf('store.h5','table',append=True) In [54]: read_hdf('store.h5', 'table', where = ['index>2']) Out[54]: A B 3 3 3 4 4 4 [2 rows x 2 columns]provide dotted attribute access to
getfrom stores, e.g.store.df == store['df']new keywords
iterator=boolean, andchunksize=number_in_a_chunkare provided to support iteration onselectandselect_as_multiple(GH3076)You can now select timestamps from an unordered timeseries similarly to an ordered timeseries (GH2437)
You can now select with a string from a DataFrame with a datelike index, in a similar way to a Series (GH3070)
In [55]: idx = date_range("2001-10-1", periods=5, freq='M') In [56]: ts = Series(np.random.rand(len(idx)),index=idx) In [57]: ts['2001'] Out[57]: 2001-10-31 0.663256 2001-11-30 0.079126 2001-12-31 0.587699 Freq: M, Length: 3, dtype: float64 In [58]: df = DataFrame(dict(A = ts)) In [59]: df['2001'] Out[59]: A 2001-10-31 0.663256 2001-11-30 0.079126 2001-12-31 0.587699 [3 rows x 1 columns]
Squeezeto possibly remove length 1 dimensions from an object.In [60]: p = Panel(randn(3,4,4),items=['ItemA','ItemB','ItemC'], ....: major_axis=date_range('20010102',periods=4), ....: minor_axis=['A','B','C','D']) ....: In [61]: p Out[61]: <class 'pandas.core.panel.Panel'> Dimensions: 3 (items) x 4 (major_axis) x 4 (minor_axis) Items axis: ItemA to ItemC Major_axis axis: 2001-01-02 00:00:00 to 2001-01-05 00:00:00 Minor_axis axis: A to D In [62]: p.reindex(items=['ItemA']).squeeze() �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[62]: A B C D 2001-01-02 -1.203403 0.425882 -0.436045 -0.982462 2001-01-03 0.348090 -0.969649 0.121731 0.202798 2001-01-04 1.215695 -0.218549 -0.631381 -0.337116 2001-01-05 0.404238 0.907213 -0.865657 0.483186 [4 rows x 4 columns] In [63]: p.reindex(items=['ItemA'],minor=['B']).squeeze() ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[63]: 2001-01-02 0.425882 2001-01-03 -0.969649 2001-01-04 -0.218549 2001-01-05 0.907213 Freq: D, Name: B, Length: 4, dtype: float64In
pd.io.data.Options,
- Fix bug when trying to fetch data for the current month when already past expiry.
- Now using lxml to scrape html instead of BeautifulSoup (lxml was faster).
- New instance variables for calls and puts are automatically created when a method that creates them is called. This works for current month where the instance variables are simply
callsandputs. Also works for future expiry months and save the instance variable ascallsMMYYorputsMMYY, whereMMYYare, respectively, the month and year of the option’s expiry.Options.get_near_stock_pricenow allows the user to specify the month for which to get relevant options data.Options.get_forward_datanow has optional kwargsnearandabove_below. This allows the user to specify if they would like to only return forward looking data for options near the current stock price. This just obtains the data from Options.get_near_stock_price instead of Options.get_xxx_data() (GH2758).Cursor coordinate information is now displayed in time-series plots.
added option display.max_seq_items to control the number of elements printed per sequence pprinting it. (GH2979)
added option display.chop_threshold to control display of small numerical values. (GH2739)
added option display.max_info_rows to prevent verbose_info from being calculated for frames above 1M rows (configurable). (GH2807, GH2918)
value_counts() now accepts a “normalize” argument, for normalized histograms. (GH2710).
DataFrame.from_records now accepts not only dicts but any instance of the collections.Mapping ABC.
added option display.mpl_style providing a sleeker visual style for plots. Based on https://gist.github.com/huyng/816622 (GH3075).
Treat boolean values as integers (values 1 and 0) for numeric operations. (GH2641)
to_html() now accepts an optional “escape” argument to control reserved HTML character escaping (enabled by default) and escapes
&, in addition to<and>. (GH2919)
See the full release notes or issue tracker on GitHub for a complete list.
v0.10.1 (January 22, 2013)¶
This is a minor release from 0.10.0 and includes new features, enhancements, and bug fixes. In particular, there is substantial new HDFStore functionality contributed by Jeff Reback.
An undesired API breakage with functions taking the inplace option has been
reverted and deprecation warnings added.
API changes¶
- Functions taking an
inplaceoption return the calling object as before. A deprecation message has been added - Groupby aggregations Max/Min no longer exclude non-numeric data (GH2700)
- Resampling an empty DataFrame now returns an empty DataFrame instead of raising an exception (GH2640)
- The file reader will now raise an exception when NA values are found in an explicitly specified integer column instead of converting the column to float (GH2631)
- DatetimeIndex.unique now returns a DatetimeIndex with the same name and
- timezone instead of an array (GH2563)
New features¶
- MySQL support for database (contribution from Dan Allan)
HDFStore¶
You may need to upgrade your existing data files. Please visit the compatibility section in the main docs.
You can designate (and index) certain columns that you want to be able to
perform queries on a table, by passing a list to data_columns
In [1]: store = HDFStore('store.h5')
In [2]: df = DataFrame(randn(8, 3), index=date_range('1/1/2000', periods=8),
...: columns=['A', 'B', 'C'])
...:
In [3]: df['string'] = 'foo'
In [4]: df.loc[df.index[4:6], 'string'] = np.nan
In [5]: df.loc[df.index[7:9], 'string'] = 'bar'
In [6]: df['string2'] = 'cool'
In [7]: df
Out[7]:
A B C string string2
2000-01-01 1.885136 -0.183873 2.550850 foo cool
2000-01-02 0.180759 -1.117089 0.061462 foo cool
2000-01-03 -0.294467 -0.591411 -0.876691 foo cool
2000-01-04 3.127110 1.451130 0.045152 foo cool
2000-01-05 -0.242846 1.195819 1.533294 NaN cool
2000-01-06 0.820521 -0.281201 1.651561 NaN cool
2000-01-07 -0.034086 0.252394 -0.498772 foo cool
2000-01-08 -2.290958 -1.601262 -0.256718 bar cool
[8 rows x 5 columns]
# on-disk operations
In [8]: store.append('df', df, data_columns = ['B','C','string','string2'])
In [9]: store.select('df', "B>0 and string=='foo'")
Out[9]:
A B C string string2
2000-01-04 3.127110 1.451130 0.045152 foo cool
2000-01-07 -0.034086 0.252394 -0.498772 foo cool
[2 rows x 5 columns]
# this is in-memory version of this type of selection
In [10]: df[(df.B > 0) & (df.string == 'foo')]
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[10]:
A B C string string2
2000-01-04 3.127110 1.451130 0.045152 foo cool
2000-01-07 -0.034086 0.252394 -0.498772 foo cool
[2 rows x 5 columns]
Retrieving unique values in an indexable or data column.
# note that this is deprecated as of 0.14.0
# can be replicated by: store.select_column('df','index').unique()
store.unique('df','index')
store.unique('df','string')
You can now store datetime64 in data columns
In [11]: df_mixed = df.copy()
In [12]: df_mixed['datetime64'] = Timestamp('20010102')
In [13]: df_mixed.loc[df_mixed.index[3:4], ['A','B']] = np.nan
In [14]: store.append('df_mixed', df_mixed)
In [15]: df_mixed1 = store.select('df_mixed')
In [16]: df_mixed1
Out[16]:
A B C string string2 datetime64
2000-01-01 1.885136 -0.183873 2.550850 foo cool 2001-01-02
2000-01-02 0.180759 -1.117089 0.061462 foo cool 2001-01-02
2000-01-03 -0.294467 -0.591411 -0.876691 foo cool 2001-01-02
2000-01-04 NaN NaN 0.045152 foo cool 2001-01-02
2000-01-05 -0.242846 1.195819 1.533294 NaN cool 2001-01-02
2000-01-06 0.820521 -0.281201 1.651561 NaN cool 2001-01-02
2000-01-07 -0.034086 0.252394 -0.498772 foo cool 2001-01-02
2000-01-08 -2.290958 -1.601262 -0.256718 bar cool 2001-01-02
[8 rows x 6 columns]
In [17]: df_mixed1.get_dtype_counts()
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[17]:
datetime64[ns] 1
float64 3
object 2
Length: 3, dtype: int64
You can pass columns keyword to select to filter a list of the return
columns, this is equivalent to passing a
Term('columns',list_of_columns_to_filter)
In [18]: store.select('df',columns = ['A','B'])
Out[18]:
A B
2000-01-01 1.885136 -0.183873
2000-01-02 0.180759 -1.117089
2000-01-03 -0.294467 -0.591411
2000-01-04 3.127110 1.451130
2000-01-05 -0.242846 1.195819
2000-01-06 0.820521 -0.281201
2000-01-07 -0.034086 0.252394
2000-01-08 -2.290958 -1.601262
[8 rows x 2 columns]
HDFStore now serializes multi-index dataframes when appending tables.
In [19]: index = MultiIndex(levels=[['foo', 'bar', 'baz', 'qux'],
....: ['one', 'two', 'three']],
....: labels=[[0, 0, 0, 1, 1, 2, 2, 3, 3, 3],
....: [0, 1, 2, 0, 1, 1, 2, 0, 1, 2]],
....: names=['foo', 'bar'])
....:
In [20]: df = DataFrame(np.random.randn(10, 3), index=index,
....: columns=['A', 'B', 'C'])
....:
In [21]: df
Out[21]:
A B C
foo bar
foo one 0.239369 0.174122 -1.131794
two -1.948006 0.980347 -0.674429
three -0.361633 -0.761218 1.768215
bar one 0.152288 -0.862613 -0.210968
two -0.859278 1.498195 0.462413
baz two -0.647604 1.511487 -0.727189
three -0.342928 -0.007364 1.427674
qux one 0.104020 2.052171 -1.230963
two -0.019240 -1.713238 0.838912
three -0.637855 0.215109 -1.515362
[10 rows x 3 columns]
In [22]: store.append('mi',df)
In [23]: store.select('mi')
Out[23]:
A B C
foo bar
foo one 0.239369 0.174122 -1.131794
two -1.948006 0.980347 -0.674429
three -0.361633 -0.761218 1.768215
bar one 0.152288 -0.862613 -0.210968
two -0.859278 1.498195 0.462413
baz two -0.647604 1.511487 -0.727189
three -0.342928 -0.007364 1.427674
qux one 0.104020 2.052171 -1.230963
two -0.019240 -1.713238 0.838912
three -0.637855 0.215109 -1.515362
[10 rows x 3 columns]
# the levels are automatically included as data columns
In [24]: store.select('mi', "foo='bar'")
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[24]:
A B C
foo bar
bar one 0.152288 -0.862613 -0.210968
two -0.859278 1.498195 0.462413
[2 rows x 3 columns]
Multi-table creation via append_to_multiple and selection via
select_as_multiple can create/select from multiple tables and return a
combined result, by using where on a selector table.
In [25]: df_mt = DataFrame(randn(8, 6), index=date_range('1/1/2000', periods=8),
....: columns=['A', 'B', 'C', 'D', 'E', 'F'])
....:
In [26]: df_mt['foo'] = 'bar'
# you can also create the tables individually
In [27]: store.append_to_multiple({ 'df1_mt' : ['A','B'], 'df2_mt' : None }, df_mt, selector = 'df1_mt')
In [28]: store
Out[28]:
<class 'pandas.io.pytables.HDFStore'>
File path: store.h5
/df frame_table (typ->appendable,nrows->8,ncols->5,indexers->[index],dc->[B,C,string,string2])
/df1_mt frame_table (typ->appendable,nrows->8,ncols->2,indexers->[index],dc->[A,B])
/df2_mt frame_table (typ->appendable,nrows->8,ncols->5,indexers->[index])
/df_mixed frame_table (typ->appendable,nrows->8,ncols->6,indexers->[index])
/mi frame_table (typ->appendable_multi,nrows->10,ncols->5,indexers->[index],dc->[bar,foo])
# indiviual tables were created
In [29]: store.select('df1_mt')
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[29]:
A B
2000-01-01 1.586924 -0.447974
2000-01-02 -0.102206 0.870302
2000-01-03 1.249874 1.458210
2000-01-04 -0.616293 0.150468
2000-01-05 -0.431163 0.016640
2000-01-06 0.800353 -0.451572
2000-01-07 1.239198 0.185437
2000-01-08 -0.040863 0.290110
[8 rows x 2 columns]
In [30]: store.select('df2_mt')
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[30]:
C D E F foo
2000-01-01 -1.573998 0.630925 -0.071659 -1.277640 bar
2000-01-02 1.275280 -1.199212 1.060780 1.673018 bar
2000-01-03 -0.710542 0.825392 1.557329 1.993441 bar
2000-01-04 0.132104 0.580923 -0.128750 1.445964 bar
2000-01-05 0.904578 -1.645852 -0.688741 0.228006 bar
2000-01-06 0.831767 0.228760 0.932498 -2.200069 bar
2000-01-07 -0.540770 -0.370038 1.298390 1.662964 bar
2000-01-08 -0.096145 1.717830 -0.462446 -0.112019 bar
[8 rows x 5 columns]
# as a multiple
In [31]: store.select_as_multiple(['df1_mt','df2_mt'], where = [ 'A>0','B>0' ], selector = 'df1_mt')
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[31]:
A B C D E F foo
2000-01-03 1.249874 1.458210 -0.710542 0.825392 1.557329 1.993441 bar
2000-01-07 1.239198 0.185437 -0.540770 -0.370038 1.298390 1.662964 bar
[2 rows x 7 columns]
Enhancements
HDFStorenow can read native PyTables table format tables- You can pass
nan_rep = 'my_nan_rep'to append, to change the default nan representation on disk (which converts to/from np.nan), this defaults to nan. - You can pass
indextoappend. This defaults toTrue. This will automagically create indicies on the indexables and data columns of the table - You can pass
chunksize=an integertoappend, to change the writing chunksize (default is 50000). This will signficantly lower your memory usage on writing. - You can pass
expectedrows=an integerto the firstappend, to set the TOTAL number of expectedrows thatPyTableswill expected. This will optimize read/write performance. Selectnow supports passingstartandstopto provide selection space limiting in selection.- Greatly improved ISO8601 (e.g., yyyy-mm-dd) date parsing for file parsers (GH2698)
- Allow
DataFrame.mergeto handle combinatorial sizes too large for 64-bit integer (GH2690) - Series now has unary negation (-series) and inversion (~series) operators (GH2686)
- DataFrame.plot now includes a
logxparameter to change the x-axis to log scale (GH2327) - Series arithmetic operators can now handle constant and ndarray input (GH2574)
- ExcelFile now takes a
kindargument to specify the file type (GH2613) - A faster implementation for Series.str methods (GH2602)
Bug Fixes
HDFStoretables can now storefloat32types correctly (cannot be mixed withfloat64however)- Fixed Google Analytics prefix when specifying request segment (GH2713).
- Function to reset Google Analytics token store so users can recover from improperly setup client secrets (GH2687).
- Fixed groupby bug resulting in segfault when passing in MultiIndex (GH2706)
- Fixed bug where passing a Series with datetime64 values into to_datetime results in bogus output values (GH2699)
- Fixed bug in
pattern in HDFStoreexpressions when pattern is not a valid regex (GH2694) - Fixed performance issues while aggregating boolean data (GH2692)
- When given a boolean mask key and a Series of new values, Series __setitem__ will now align the incoming values with the original Series (GH2686)
- Fixed MemoryError caused by performing counting sort on sorting MultiIndex levels with a very large number of combinatorial values (GH2684)
- Fixed bug that causes plotting to fail when the index is a DatetimeIndex with a fixed-offset timezone (GH2683)
- Corrected businessday subtraction logic when the offset is more than 5 bdays and the starting date is on a weekend (GH2680)
- Fixed C file parser behavior when the file has more columns than data (GH2668)
- Fixed file reader bug that misaligned columns with data in the presence of an implicit column and a specified usecols value
- DataFrames with numerical or datetime indices are now sorted prior to plotting (GH2609)
- Fixed DataFrame.from_records error when passed columns, index, but empty records (GH2633)
- Several bug fixed for Series operations when dtype is datetime64 (GH2689, GH2629, GH2626)
See the full release notes or issue tracker on GitHub for a complete list.
v0.10.0 (December 17, 2012)¶
This is a major release from 0.9.1 and includes many new features and enhancements along with a large number of bug fixes. There are also a number of important API changes that long-time pandas users should pay close attention to.
File parsing new features¶
The delimited file parsing engine (the guts of read_csv and read_table)
has been rewritten from the ground up and now uses a fraction the amount of
memory while parsing, while being 40% or more faster in most use cases (in some
cases much faster).
There are also many new features:
- Much-improved Unicode handling via the
encodingoption. - Column filtering (
usecols) - Dtype specification (
dtypeargument) - Ability to specify strings to be recognized as True/False
- Ability to yield NumPy record arrays (
as_recarray) - High performance
delim_whitespaceoption - Decimal format (e.g. European format) specification
- Easier CSV dialect options:
escapechar,lineterminator,quotechar, etc. - More robust handling of many exceptional kinds of files observed in the wild
API changes¶
Deprecated DataFrame BINOP TimeSeries special case behavior
The default behavior of binary operations between a DataFrame and a Series has always been to align on the DataFrame’s columns and broadcast down the rows, except in the special case that the DataFrame contains time series. Since there are now method for each binary operator enabling you to specify how you want to broadcast, we are phasing out this special case (Zen of Python: Special cases aren’t special enough to break the rules). Here’s what I’m talking about:
In [1]: import pandas as pd
In [2]: df = pd.DataFrame(np.random.randn(6, 4),
...: index=pd.date_range('1/1/2000', periods=6))
...:
In [3]: df
Out[3]:
0 1 2 3
2000-01-01 -0.134024 -0.205969 1.348944 -1.198246
2000-01-02 -1.626124 0.982041 0.059493 -0.460111
2000-01-03 -1.565401 -0.025706 0.942864 2.502156
2000-01-04 -0.302741 0.261551 -0.066342 0.897097
2000-01-05 0.268766 -1.225092 0.582752 -1.490764
2000-01-06 -0.639757 -0.952750 -0.892402 0.505987
[6 rows x 4 columns]
# deprecated now
In [4]: df - df[0]
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[4]:
2000-01-01 00:00:00 2000-01-02 00:00:00 2000-01-03 00:00:00 \
2000-01-01 NaN NaN NaN
2000-01-02 NaN NaN NaN
2000-01-03 NaN NaN NaN
2000-01-04 NaN NaN NaN
2000-01-05 NaN NaN NaN
2000-01-06 NaN NaN NaN
2000-01-04 00:00:00 2000-01-05 00:00:00 2000-01-06 00:00:00 0 \
2000-01-01 NaN NaN NaN NaN
2000-01-02 NaN NaN NaN NaN
2000-01-03 NaN NaN NaN NaN
2000-01-04 NaN NaN NaN NaN
2000-01-05 NaN NaN NaN NaN
2000-01-06 NaN NaN NaN NaN
1 2 3
2000-01-01 NaN NaN NaN
2000-01-02 NaN NaN NaN
2000-01-03 NaN NaN NaN
2000-01-04 NaN NaN NaN
2000-01-05 NaN NaN NaN
2000-01-06 NaN NaN NaN
[6 rows x 10 columns]
# Change your code to
In [5]: df.sub(df[0], axis=0) # align on axis 0 (rows)
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[5]:
0 1 2 3
2000-01-01 0.0 -0.071946 1.482967 -1.064223
2000-01-02 0.0 2.608165 1.685618 1.166013
2000-01-03 0.0 1.539695 2.508265 4.067556
2000-01-04 0.0 0.564293 0.236399 1.199839
2000-01-05 0.0 -1.493857 0.313986 -1.759530
2000-01-06 0.0 -0.312993 -0.252645 1.145744
[6 rows x 4 columns]
You will get a deprecation warning in the 0.10.x series, and the deprecated functionality will be removed in 0.11 or later.
Altered resample default behavior
The default time series resample binning behavior of daily D and
higher frequencies has been changed to closed='left', label='left'. Lower
nfrequencies are unaffected. The prior defaults were causing a great deal of
confusion for users, especially resampling data to daily frequency (which
labeled the aggregated group with the end of the interval: the next day).
In [1]: dates = pd.date_range('1/1/2000', '1/5/2000', freq='4h')
In [2]: series = Series(np.arange(len(dates)), index=dates)
In [3]: series
Out[3]:
2000-01-01 00:00:00 0
2000-01-01 04:00:00 1
2000-01-01 08:00:00 2
2000-01-01 12:00:00 3
2000-01-01 16:00:00 4
2000-01-01 20:00:00 5
2000-01-02 00:00:00 6
2000-01-02 04:00:00 7
2000-01-02 08:00:00 8
2000-01-02 12:00:00 9
2000-01-02 16:00:00 10
2000-01-02 20:00:00 11
2000-01-03 00:00:00 12
2000-01-03 04:00:00 13
2000-01-03 08:00:00 14
2000-01-03 12:00:00 15
2000-01-03 16:00:00 16
2000-01-03 20:00:00 17
2000-01-04 00:00:00 18
2000-01-04 04:00:00 19
2000-01-04 08:00:00 20
2000-01-04 12:00:00 21
2000-01-04 16:00:00 22
2000-01-04 20:00:00 23
2000-01-05 00:00:00 24
Freq: 4H, dtype: int64
In [4]: series.resample('D', how='sum')
Out[4]:
2000-01-01 15
2000-01-02 51
2000-01-03 87
2000-01-04 123
2000-01-05 24
Freq: D, dtype: int64
In [5]: # old behavior
In [6]: series.resample('D', how='sum', closed='right', label='right')
Out[6]:
2000-01-01 0
2000-01-02 21
2000-01-03 57
2000-01-04 93
2000-01-05 129
Freq: D, dtype: int64
- Infinity and negative infinity are no longer treated as NA by
isnullandnotnull. That they ever were was a relic of early pandas. This behavior can be re-enabled globally by themode.use_inf_as_nulloption:
In [6]: s = pd.Series([1.5, np.inf, 3.4, -np.inf])
In [7]: pd.isnull(s)
Out[7]:
0 False
1 False
2 False
3 False
Length: 4, dtype: bool
In [8]: s.fillna(0)
����������������������������������������������������������������������������Out[8]:
0 1.500000
1 inf
2 3.400000
3 -inf
Length: 4, dtype: float64
In [9]: pd.set_option('use_inf_as_null', True)
In [10]: pd.isnull(s)
Out[10]:
0 False
1 True
2 False
3 True
Length: 4, dtype: bool
In [11]: s.fillna(0)
�����������������������������������������������������������������������������Out[11]:
0 1.5
1 0.0
2 3.4
3 0.0
Length: 4, dtype: float64
In [12]: pd.reset_option('use_inf_as_null')
- Methods with the
inplaceoption now all returnNoneinstead of the calling object. E.g. code written likedf = df.fillna(0, inplace=True)may stop working. To fix, simply delete the unnecessary variable assignment. pandas.mergeno longer sorts the group keys (sort=False) by default. This was done for performance reasons: the group-key sorting is often one of the more expensive parts of the computation and is often unnecessary.- The default column names for a file with no header have been changed to the
integers
0throughN - 1. This is to create consistency with the DataFrame constructor with no columns specified. The v0.9.0 behavior (namesX0,X1, ...) can be reproduced by specifyingprefix='X':
In [13]: data= 'a,b,c\n1,Yes,2\n3,No,4'
In [14]: print(data)
a,b,c
1,Yes,2
3,No,4
In [15]: pd.read_csv(StringIO(data), header=None)
���������������������Out[15]:
0 1 2
0 a b c
1 1 Yes 2
2 3 No 4
[3 rows x 3 columns]
In [16]: pd.read_csv(StringIO(data), header=None, prefix='X')
���������������������������������������������������������������������������������������������������������Out[16]:
X0 X1 X2
0 a b c
1 1 Yes 2
2 3 No 4
[3 rows x 3 columns]
- Values like
'Yes'and'No'are not interpreted as boolean by default, though this can be controlled by newtrue_valuesandfalse_valuesarguments:
In [17]: print(data)
a,b,c
1,Yes,2
3,No,4
In [18]: pd.read_csv(StringIO(data))
���������������������Out[18]:
a b c
0 1 Yes 2
1 3 No 4
[2 rows x 3 columns]
In [19]: pd.read_csv(StringIO(data), true_values=['Yes'], false_values=['No'])
��������������������������������������������������������������������������������������������Out[19]:
a b c
0 1 True 2
1 3 False 4
[2 rows x 3 columns]
- The file parsers will not recognize non-string values arising from a
converter function as NA if passed in the
na_valuesargument. It’s better to do post-processing using thereplacefunction instead. - Calling
fillnaon Series or DataFrame with no arguments is no longer valid code. You must either specify a fill value or an interpolation method:
In [20]: s = Series([np.nan, 1., 2., np.nan, 4])
In [21]: s
Out[21]:
0 NaN
1 1.0
2 2.0
3 NaN
4 4.0
Length: 5, dtype: float64
In [22]: s.fillna(0)
���������������������������������������������������������������������������������Out[22]:
0 0.0
1 1.0
2 2.0
3 0.0
4 4.0
Length: 5, dtype: float64
In [23]: s.fillna(method='pad')
������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[23]:
0 NaN
1 1.0
2 2.0
3 2.0
4 4.0
Length: 5, dtype: float64
Convenience methods ffill and bfill have been added:
In [24]: s.ffill()
Out[24]:
0 NaN
1 1.0
2 2.0
3 2.0
4 4.0
Length: 5, dtype: float64
Series.applywill now operate on a returned value from the applied function, that is itself a series, and possibly upcast the result to a DataFrameIn [25]: def f(x): ....: return Series([ x, x**2 ], index = ['x', 'x^2']) ....: In [26]: s = Series(np.random.rand(5)) In [27]: s Out[27]: 0 0.717478 1 0.815199 2 0.452478 3 0.848385 4 0.235477 Length: 5, dtype: float64 In [28]: s.apply(f) ����������������������������������������������������������������������������������������������������������Out[28]: x x^2 0 0.717478 0.514775 1 0.815199 0.664550 2 0.452478 0.204737 3 0.848385 0.719757 4 0.235477 0.055449 [5 rows x 2 columns]
New API functions for working with pandas options (GH2097):
get_option/set_option- get/set the value of an option. Partial names are accepted. -reset_option- reset one or more options to their default value. Partial names are accepted. -describe_option- print a description of one or more options. When called with no arguments. print all registered options.
Note:
set_printoptions/reset_printoptionsare now deprecated (but functioning), the print options now live under “display.XYZ”. For example:In [29]: get_option("display.max_rows") Out[29]: 15
to_string() methods now always return unicode strings (GH2224).
New features¶
Wide DataFrame Printing¶
Instead of printing the summary information, pandas now splits the string representation across multiple rows by default:
In [30]: wide_frame = DataFrame(randn(5, 16))
In [31]: wide_frame
Out[31]:
0 1 2 3 4 5 6 \
0 -0.681624 0.191356 1.180274 -0.834179 0.703043 0.166568 -0.583599
1 0.441522 -0.316864 -0.017062 1.570114 -0.360875 -0.880096 0.235532
2 -0.412451 -0.462580 0.422194 0.288403 -0.487393 -0.777639 0.055865
3 -0.277255 1.331263 0.585174 -0.568825 -0.719412 1.191340 -0.456362
4 -1.642511 0.432560 1.218080 -0.564705 -0.581790 0.286071 0.048725
7 8 9 10 11 12 13 \
0 -1.201796 -1.422811 -0.882554 1.209871 -0.941235 0.863067 -0.336232
1 0.207232 -1.983857 -1.702547 -1.621234 -0.906840 1.014601 -0.475108
2 1.383381 0.085638 0.246392 0.965887 0.246354 -0.727728 -0.094414
3 0.089931 0.776079 0.752889 -1.195795 -1.425911 -0.548829 0.774225
4 1.002440 1.276582 0.054399 0.241963 -0.471786 0.314510 -0.059986
14 15
0 -0.976847 0.033862
1 -0.358944 1.262942
2 -0.276854 0.158399
3 0.740501 1.510263
4 -2.069319 -1.115104
[5 rows x 16 columns]
The old behavior of printing out summary information can be achieved via the ‘expand_frame_repr’ print option:
In [32]: pd.set_option('expand_frame_repr', False)
In [33]: wide_frame
Out[33]:
0 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15
0 -0.681624 0.191356 1.180274 -0.834179 0.703043 0.166568 -0.583599 -1.201796 -1.422811 -0.882554 1.209871 -0.941235 0.863067 -0.336232 -0.976847 0.033862
1 0.441522 -0.316864 -0.017062 1.570114 -0.360875 -0.880096 0.235532 0.207232 -1.983857 -1.702547 -1.621234 -0.906840 1.014601 -0.475108 -0.358944 1.262942
2 -0.412451 -0.462580 0.422194 0.288403 -0.487393 -0.777639 0.055865 1.383381 0.085638 0.246392 0.965887 0.246354 -0.727728 -0.094414 -0.276854 0.158399
3 -0.277255 1.331263 0.585174 -0.568825 -0.719412 1.191340 -0.456362 0.089931 0.776079 0.752889 -1.195795 -1.425911 -0.548829 0.774225 0.740501 1.510263
4 -1.642511 0.432560 1.218080 -0.564705 -0.581790 0.286071 0.048725 1.002440 1.276582 0.054399 0.241963 -0.471786 0.314510 -0.059986 -2.069319 -1.115104
[5 rows x 16 columns]
The width of each line can be changed via ‘line_width’ (80 by default):
In [34]: pd.set_option('line_width', 40)
line_width has been deprecated, use display.width instead (currently both are
identical)
In [35]: wide_frame
������������������������������������������������������������������������������������������Out[35]:
0 1 2 \
0 -0.681624 0.191356 1.180274
1 0.441522 -0.316864 -0.017062
2 -0.412451 -0.462580 0.422194
3 -0.277255 1.331263 0.585174
4 -1.642511 0.432560 1.218080
3 4 5 \
0 -0.834179 0.703043 0.166568
1 1.570114 -0.360875 -0.880096
2 0.288403 -0.487393 -0.777639
3 -0.568825 -0.719412 1.191340
4 -0.564705 -0.581790 0.286071
6 7 8 \
0 -0.583599 -1.201796 -1.422811
1 0.235532 0.207232 -1.983857
2 0.055865 1.383381 0.085638
3 -0.456362 0.089931 0.776079
4 0.048725 1.002440 1.276582
9 10 11 \
0 -0.882554 1.209871 -0.941235
1 -1.702547 -1.621234 -0.906840
2 0.246392 0.965887 0.246354
3 0.752889 -1.195795 -1.425911
4 0.054399 0.241963 -0.471786
12 13 14 \
0 0.863067 -0.336232 -0.976847
1 1.014601 -0.475108 -0.358944
2 -0.727728 -0.094414 -0.276854
3 -0.548829 0.774225 0.740501
4 0.314510 -0.059986 -2.069319
15
0 0.033862
1 1.262942
2 0.158399
3 1.510263
4 -1.115104
[5 rows x 16 columns]
Updated PyTables Support¶
Docs for PyTables Table format & several enhancements to the api. Here is a taste of what to expect.
In [36]: store = HDFStore('store.h5')
In [37]: df = DataFrame(randn(8, 3), index=date_range('1/1/2000', periods=8),
....: columns=['A', 'B', 'C'])
....:
In [38]: df
Out[38]:
A B C
2000-01-01 -0.369325 -1.502617 -0.376280
2000-01-02 0.511936 -0.116412 -0.625256
2000-01-03 -0.550627 1.261433 -0.552429
2000-01-04 1.695803 -1.025917 -0.910942
2000-01-05 0.426805 -0.131749 0.432600
2000-01-06 0.044671 -0.341265 1.844536
2000-01-07 -2.036047 0.000830 -0.955697
2000-01-08 -0.898872 -0.725411 0.059904
[8 rows x 3 columns]
# appending data frames
In [39]: df1 = df[0:4]
In [40]: df2 = df[4:]
In [41]: store.append('df', df1)
In [42]: store.append('df', df2)
In [43]: store
Out[43]:
<class 'pandas.io.pytables.HDFStore'>
File path: store.h5
/df frame_table (typ->appendable,nrows->8,ncols->3,indexers->[index])
# selecting the entire store
In [44]: store.select('df')
������������������������������������������������������������������������������������������������������������������������������������������������������Out[44]:
A B C
2000-01-01 -0.369325 -1.502617 -0.376280
2000-01-02 0.511936 -0.116412 -0.625256
2000-01-03 -0.550627 1.261433 -0.552429
2000-01-04 1.695803 -1.025917 -0.910942
2000-01-05 0.426805 -0.131749 0.432600
2000-01-06 0.044671 -0.341265 1.844536
2000-01-07 -2.036047 0.000830 -0.955697
2000-01-08 -0.898872 -0.725411 0.059904
[8 rows x 3 columns]
In [45]: wp = Panel(randn(2, 5, 4), items=['Item1', 'Item2'],
....: major_axis=date_range('1/1/2000', periods=5),
....: minor_axis=['A', 'B', 'C', 'D'])
....:
In [46]: wp
Out[46]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 5 (major_axis) x 4 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 2000-01-01 00:00:00 to 2000-01-05 00:00:00
Minor_axis axis: A to D
# storing a panel
In [47]: store.append('wp',wp)
# selecting via A QUERY
In [48]: store.select('wp', "major_axis>20000102 and minor_axis=['A','B']")
Out[48]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 3 (major_axis) x 2 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 2000-01-03 00:00:00 to 2000-01-05 00:00:00
Minor_axis axis: A to B
# removing data from tables
In [49]: store.remove('wp', "major_axis>20000103")
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[49]: 8
In [50]: store.select('wp')
������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[50]:
<class 'pandas.core.panel.Panel'>
Dimensions: 2 (items) x 3 (major_axis) x 4 (minor_axis)
Items axis: Item1 to Item2
Major_axis axis: 2000-01-01 00:00:00 to 2000-01-03 00:00:00
Minor_axis axis: A to D
# deleting a store
In [51]: del store['df']
In [52]: store
Out[52]:
<class 'pandas.io.pytables.HDFStore'>
File path: store.h5
/wp wide_table (typ->appendable,nrows->12,ncols->2,indexers->[major_axis,minor_axis])
Enhancements
added ability to hierarchical keys
In [53]: store.put('foo/bar/bah', df) In [54]: store.append('food/orange', df) In [55]: store.append('food/apple', df) In [56]: store Out[56]: <class 'pandas.io.pytables.HDFStore'> File path: store.h5 /foo/bar/bah frame (shape->[8,3]) /food/apple frame_table (typ->appendable,nrows->8,ncols->3,indexers->[index]) /food/orange frame_table (typ->appendable,nrows->8,ncols->3,indexers->[index]) /wp wide_table (typ->appendable,nrows->12,ncols->2,indexers->[major_axis,minor_axis]) # remove all nodes under this level In [57]: store.remove('food') In [58]: store Out[58]: <class 'pandas.io.pytables.HDFStore'> File path: store.h5 /foo/bar/bah frame (shape->[8,3]) /wp wide_table (typ->appendable,nrows->12,ncols->2,indexers->[major_axis,minor_axis])
added mixed-dtype support!
In [59]: df['string'] = 'string' In [60]: df['int'] = 1 In [61]: store.append('df',df) In [62]: df1 = store.select('df') In [63]: df1 Out[63]: A B C string int 2000-01-01 -0.369325 -1.502617 -0.376280 string 1 2000-01-02 0.511936 -0.116412 -0.625256 string 1 2000-01-03 -0.550627 1.261433 -0.552429 string 1 2000-01-04 1.695803 -1.025917 -0.910942 string 1 2000-01-05 0.426805 -0.131749 0.432600 string 1 2000-01-06 0.044671 -0.341265 1.844536 string 1 2000-01-07 -2.036047 0.000830 -0.955697 string 1 2000-01-08 -0.898872 -0.725411 0.059904 string 1 [8 rows x 5 columns] In [64]: df1.get_dtype_counts() ��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[64]: float64 3 int64 1 object 1 Length: 3, dtype: int64
performance improvments on table writing
support for arbitrarily indexed dimensions
SparseSeriesnow has adensityproperty (GH2384)enable
Series.str.strip/lstrip/rstripmethods to take an input argument to strip arbitrary characters (GH2411)implement
value_varsinmeltto limit values to certain columns and addmeltto pandas namespace (GH2412)
Bug Fixes
- added
Termmethod of specifying where conditions (GH1996). del store['df']now callstore.remove('df')for store deletion- deleting of consecutive rows is much faster than before
min_itemsizeparameter can be specified in table creation to force a minimum size for indexing columns (the previous implementation would set the column size based on the first append)- indexing support via
create_table_index(requires PyTables >= 2.3) (GH698). - appending on a store would fail if the table was not first created via
put - fixed issue with missing attributes after loading a pickled dataframe (GH2431)
- minor change to select and remove: require a table ONLY if where is also provided (and not None)
Compatibility
0.10 of HDFStore is backwards compatible for reading tables created in a prior version of pandas,
however, query terms using the prior (undocumented) methodology are unsupported. You must read in the entire
file and write it out using the new format to take advantage of the updates.
N Dimensional Panels (Experimental)¶
Adding experimental support for Panel4D and factory functions to create n-dimensional named panels. Docs for NDim. Here is a taste of what to expect.
In [65]: p4d = Panel4D(randn(2, 2, 5, 4), ....: labels=['Label1','Label2'], ....: items=['Item1', 'Item2'], ....: major_axis=date_range('1/1/2000', periods=5), ....: minor_axis=['A', 'B', 'C', 'D']) ....: In [66]: p4d Out[66]: <class 'pandas.core.panelnd.Panel4D'> Dimensions: 2 (labels) x 2 (items) x 5 (major_axis) x 4 (minor_axis) Labels axis: Label1 to Label2 Items axis: Item1 to Item2 Major_axis axis: 2000-01-01 00:00:00 to 2000-01-05 00:00:00 Minor_axis axis: A to D
See the full release notes or issue tracker on GitHub for a complete list.
v0.9.1 (November 14, 2012)¶
This is a bugfix release from 0.9.0 and includes several new features and enhancements along with a large number of bug fixes. The new features include by-column sort order for DataFrame and Series, improved NA handling for the rank method, masking functions for DataFrame, and intraday time-series filtering for DataFrame.
New features¶
Series.sort, DataFrame.sort, and DataFrame.sort_index can now be specified in a per-column manner to support multiple sort orders (GH928)
In [2]: df = DataFrame(np.random.randint(0, 2, (6, 3)), columns=['A', 'B', 'C']) In [3]: df.sort(['A', 'B'], ascending=[1, 0]) Out[3]: A B C 3 0 1 1 4 0 1 1 2 0 0 1 0 1 0 0 1 1 0 0 5 1 0 0DataFrame.rank now supports additional argument values for the na_option parameter so missing values can be assigned either the largest or the smallest rank (GH1508, GH2159)
In [1]: df = DataFrame(np.random.randn(6, 3), columns=['A', 'B', 'C']) In [2]: df.loc[2:4] = np.nan In [3]: df.rank() Out[3]: A B C 0 3.0 1.0 3.0 1 2.0 2.0 1.0 2 NaN NaN NaN 3 NaN NaN NaN 4 NaN NaN NaN 5 1.0 3.0 2.0 [6 rows x 3 columns] In [4]: df.rank(na_option='top') ������������������������������������������������������������������������������������������������������������������������������������������������������Out[4]: A B C 0 6.0 4.0 6.0 1 5.0 5.0 4.0 2 2.0 2.0 2.0 3 2.0 2.0 2.0 4 2.0 2.0 2.0 5 4.0 6.0 5.0 [6 rows x 3 columns] In [5]: df.rank(na_option='bottom') ������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[5]: A B C 0 3.0 1.0 3.0 1 2.0 2.0 1.0 2 5.0 5.0 5.0 3 5.0 5.0 5.0 4 5.0 5.0 5.0 5 1.0 3.0 2.0 [6 rows x 3 columns]DataFrame has new where and mask methods to select values according to a given boolean mask (GH2109, GH2151)
DataFrame currently supports slicing via a boolean vector the same length as the DataFrame (inside the []). The returned DataFrame has the same number of columns as the original, but is sliced on its index.
In [6]: df = DataFrame(np.random.randn(5, 3), columns = ['A','B','C']) In [7]: df Out[7]: A B C 0 1.744738 -0.356939 0.092791 1 1.222637 1.909179 0.195946 2 0.481559 -0.404023 -1.115882 3 2.093925 0.010808 -1.775758 4 1.303175 0.025683 -1.795489 [5 rows x 3 columns] In [8]: df[df['A'] > 0] �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[8]: A B C 0 1.744738 -0.356939 0.092791 1 1.222637 1.909179 0.195946 2 0.481559 -0.404023 -1.115882 3 2.093925 0.010808 -1.775758 4 1.303175 0.025683 -1.795489 [5 rows x 3 columns]If a DataFrame is sliced with a DataFrame based boolean condition (with the same size as the original DataFrame), then a DataFrame the same size (index and columns) as the original is returned, with elements that do not meet the boolean condition as NaN. This is accomplished via the new method DataFrame.where. In addition, where takes an optional other argument for replacement.
In [9]: df[df>0] Out[9]: A B C 0 1.744738 NaN 0.092791 1 1.222637 1.909179 0.195946 2 0.481559 NaN NaN 3 2.093925 0.010808 NaN 4 1.303175 0.025683 NaN [5 rows x 3 columns] In [10]: df.where(df>0) �������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[10]: A B C 0 1.744738 NaN 0.092791 1 1.222637 1.909179 0.195946 2 0.481559 NaN NaN 3 2.093925 0.010808 NaN 4 1.303175 0.025683 NaN [5 rows x 3 columns] In [11]: df.where(df>0,-df) ���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[11]: A B C 0 1.744738 0.356939 0.092791 1 1.222637 1.909179 0.195946 2 0.481559 0.404023 1.115882 3 2.093925 0.010808 1.775758 4 1.303175 0.025683 1.795489 [5 rows x 3 columns]Furthermore, where now aligns the input boolean condition (ndarray or DataFrame), such that partial selection with setting is possible. This is analagous to partial setting via .ix (but on the contents rather than the axis labels)
In [12]: df2 = df.copy() In [13]: df2[ df2[1:4] > 0 ] = 3 In [14]: df2 Out[14]: A B C 0 1.744738 -0.356939 0.092791 1 3.000000 3.000000 3.000000 2 3.000000 -0.404023 -1.115882 3 3.000000 3.000000 -1.775758 4 1.303175 0.025683 -1.795489 [5 rows x 3 columns]DataFrame.mask is the inverse boolean operation of where.
In [15]: df.mask(df<=0) Out[15]: A B C 0 1.744738 NaN 0.092791 1 1.222637 1.909179 0.195946 2 0.481559 NaN NaN 3 2.093925 0.010808 NaN 4 1.303175 0.025683 NaN [5 rows x 3 columns]Enable referencing of Excel columns by their column names (GH1936)
In [16]: xl = ExcelFile('data/test.xls') In [17]: xl.parse('Sheet1', index_col=0, parse_dates=True, ....: parse_cols='A:D') ....: Out[17]: A B C 2000-01-03 0.980269 3.685731 -0.364217 2000-01-04 1.047916 -0.041232 -0.161812 2000-01-05 0.498581 0.731168 -0.537677 2000-01-06 1.120202 1.567621 0.003641 2000-01-07 -0.487094 0.571455 -1.611639 2000-01-10 0.836649 0.246462 0.588543 2000-01-11 -0.157161 1.340307 1.195778 [7 rows x 3 columns]Added option to disable pandas-style tick locators and formatters using series.plot(x_compat=True) or pandas.plot_params[‘x_compat’] = True (GH2205)
Existing TimeSeries methods at_time and between_time were added to DataFrame (GH2149)
DataFrame.dot can now accept ndarrays (GH2042)
DataFrame.drop now supports non-unique indexes (GH2101)
Panel.shift now supports negative periods (GH2164)
DataFrame now support unary ~ operator (GH2110)
API changes¶
Upsampling data with a PeriodIndex will result in a higher frequency TimeSeries that spans the original time window
In [1]: prng = period_range('2012Q1', periods=2, freq='Q') In [2]: s = Series(np.random.randn(len(prng)), prng) In [4]: s.resample('M') Out[4]: 2012-01 -1.471992 2012-02 NaN 2012-03 NaN 2012-04 -0.493593 2012-05 NaN 2012-06 NaN Freq: M, dtype: float64Period.end_time now returns the last nanosecond in the time interval (GH2124, GH2125, GH1764)
In [18]: p = Period('2012') In [19]: p.end_time Out[19]: Timestamp('2012-12-31 23:59:59.999999999')File parsers no longer coerce to float or bool for columns that have custom converters specified (GH2184)
In [20]: data = 'A,B,C\n00001,001,5\n00002,002,6' In [21]: read_csv(StringIO(data), converters={'A' : lambda x: x.strip()}) Out[21]: A B C 0 00001 1 5 1 00002 2 6 [2 rows x 3 columns]
See the full release notes or issue tracker on GitHub for a complete list.
v0.9.0 (October 7, 2012)¶
This is a major release from 0.8.1 and includes several new features and enhancements along with a large number of bug fixes. New features include vectorized unicode encoding/decoding for Series.str, to_latex method to DataFrame, more flexible parsing of boolean values, and enabling the download of options data from Yahoo! Finance.
New features¶
- Add
encodeanddecodefor unicode handling to vectorized string processing methods in Series.str (GH1706)- Add
DataFrame.to_latexmethod (GH1735)- Add convenient expanding window equivalents of all rolling_* ops (GH1785)
- Add Options class to pandas.io.data for fetching options data from Yahoo! Finance (GH1748, GH1739)
- More flexible parsing of boolean values (Yes, No, TRUE, FALSE, etc) (GH1691, GH1295)
- Add
levelparameter toSeries.reset_indexTimeSeries.between_timecan now select times across midnight (GH1871)- Series constructor can now handle generator as input (GH1679)
DataFrame.dropnacan now take multiple axes (tuple/list) as input (GH924)- Enable
skip_footerparameter inExcelFile.parse(GH1843)
API changes¶
- The default column names when
header=Noneand no columns names passed to functions likeread_csvhas changed to be more Pythonic and amenable to attribute access:
In [1]: data = '0,0,1\n1,1,0\n0,1,0'
In [2]: df = read_csv(StringIO(data), header=None)
In [3]: df
Out[3]:
0 1 2
0 0 0 1
1 1 1 0
2 0 1 0
[3 rows x 3 columns]
- Creating a Series from another Series, passing an index, will cause reindexing
to happen inside rather than treating the Series like an ndarray. Technically
improper usages like
Series(df[col1], index=df[col2])that worked before “by accident” (this was never intended) will lead to all NA Series in some cases. To be perfectly clear:
In [4]: s1 = Series([1, 2, 3])
In [5]: s1
Out[5]:
0 1
1 2
2 3
Length: 3, dtype: int64
In [6]: s2 = Series(s1, index=['foo', 'bar', 'baz'])
In [7]: s2
Out[7]:
foo NaN
bar NaN
baz NaN
Length: 3, dtype: float64
- Deprecated
day_of_yearAPI removed from PeriodIndex, usedayofyear(GH1723) - Don’t modify NumPy suppress printoption to True at import time
- The internal HDF5 data arrangement for DataFrames has been transposed. Legacy files will still be readable by HDFStore (GH1834, GH1824)
- Legacy cruft removed: pandas.stats.misc.quantileTS
- Use ISO8601 format for Period repr: monthly, daily, and on down (GH1776)
- Empty DataFrame columns are now created as object dtype. This will prevent a class of TypeErrors that was occurring in code where the dtype of a column would depend on the presence of data or not (e.g. a SQL query having results) (GH1783)
- Setting parts of DataFrame/Panel using ix now aligns input Series/DataFrame (GH1630)
firstandlastmethods inGroupByno longer drop non-numeric columns (GH1809)- Resolved inconsistencies in specifying custom NA values in text parser.
na_valuesof type dict no longer override default NAs unlesskeep_default_nais set to false explicitly (GH1657) DataFrame.dotwill not do data alignment, and also work with Series (GH1915)
See the full release notes or issue tracker on GitHub for a complete list.
v0.8.1 (July 22, 2012)¶
This release includes a few new features, performance enhancements, and over 30 bug fixes from 0.8.0. New features include notably NA friendly string processing functionality and a series of new plot types and options.
New features¶
- Add vectorized string processing methods accessible via Series.str (GH620)
- Add option to disable adjustment in EWMA (GH1584)
- Radviz plot (GH1566)
- Parallel coordinates plot
- Bootstrap plot
- Per column styles and secondary y-axis plotting (GH1559)
- New datetime converters millisecond plotting (GH1599)
- Add option to disable “sparse” display of hierarchical indexes (GH1538)
- Series/DataFrame’s
set_indexmethod can append levels to an existing Index/MultiIndex (GH1569, GH1577)
Performance improvements¶
- Improved implementation of rolling min and max (thanks to Bottleneck !)
- Add accelerated
'median'GroupBy option (GH1358)- Significantly improve the performance of parsing ISO8601-format date strings with
DatetimeIndexorto_datetime(GH1571)- Improve the performance of GroupBy on single-key aggregations and use with Categorical types
- Significant datetime parsing performance improvments
v0.8.0 (June 29, 2012)¶
This is a major release from 0.7.3 and includes extensive work on the time series handling and processing infrastructure as well as a great deal of new functionality throughout the library. It includes over 700 commits from more than 20 distinct authors. Most pandas 0.7.3 and earlier users should not experience any issues upgrading, but due to the migration to the NumPy datetime64 dtype, there may be a number of bugs and incompatibilities lurking. Lingering incompatibilities will be fixed ASAP in a 0.8.1 release if necessary. See the full release notes or issue tracker on GitHub for a complete list.
Support for non-unique indexes¶
All objects can now work with non-unique indexes. Data alignment / join operations work according to SQL join semantics (including, if application, index duplication in many-to-many joins)
NumPy datetime64 dtype and 1.6 dependency¶
Time series data are now represented using NumPy’s datetime64 dtype; thus, pandas 0.8.0 now requires at least NumPy 1.6. It has been tested and verified to work with the development version (1.7+) of NumPy as well which includes some significant user-facing API changes. NumPy 1.6 also has a number of bugs having to do with nanosecond resolution data, so I recommend that you steer clear of NumPy 1.6’s datetime64 API functions (though limited as they are) and only interact with this data using the interface that pandas provides.
See the end of the 0.8.0 section for a “porting” guide listing potential issues for users migrating legacy codebases from pandas 0.7 or earlier to 0.8.0.
Bug fixes to the 0.7.x series for legacy NumPy < 1.6 users will be provided as they arise. There will be no more further development in 0.7.x beyond bug fixes.
Time series changes and improvements¶
Note
With this release, legacy scikits.timeseries users should be able to port their code to use pandas.
Note
See documentation for overview of pandas timeseries API.
- New datetime64 representation speeds up join operations and data alignment, reduces memory usage, and improve serialization / deserialization performance significantly over datetime.datetime
- High performance and flexible resample method for converting from high-to-low and low-to-high frequency. Supports interpolation, user-defined aggregation functions, and control over how the intervals and result labeling are defined. A suite of high performance Cython/C-based resampling functions (including Open-High-Low-Close) have also been implemented.
- Revamp of frequency aliases and support for frequency shortcuts like ‘15min’, or ‘1h30min’
- New DatetimeIndex class supports both fixed frequency and irregular time series. Replaces now deprecated DateRange class
- New
PeriodIndexandPeriodclasses for representing time spans and performing calendar logic, including the 12 fiscal quarterly frequencies <timeseries.quarterly>. This is a partial port of, and a substantial enhancement to, elements of the scikits.timeseries codebase. Support for conversion between PeriodIndex and DatetimeIndex - New Timestamp data type subclasses datetime.datetime, providing the same interface while enabling working with nanosecond-resolution data. Also provides easy time zone conversions.
- Enhanced support for time zones. Add
tz_convert and
tz_lcoalizemethods to TimeSeries and DataFrame. All timestamps are stored as UTC; Timestamps from DatetimeIndex objects with time zone set will be localized to localtime. Time zone conversions are therefore essentially free. User needs to know very little about pytz library now; only time zone names as as strings are required. Time zone-aware timestamps are equal if and only if their UTC timestamps match. Operations between time zone-aware time series with different time zones will result in a UTC-indexed time series. - Time series string indexing conveniences / shortcuts: slice years, year and month, and index values with strings
- Enhanced time series plotting; adaptation of scikits.timeseries matplotlib-based plotting code
- New
date_range,bdate_range, andperiod_rangefactory functions - Robust frequency inference function infer_freq and
inferred_freqproperty of DatetimeIndex, with option to infer frequency on construction of DatetimeIndex - to_datetime function efficiently parses array of strings to DatetimeIndex. DatetimeIndex will parse array or list of strings to datetime64
- Optimized support for datetime64-dtype data in Series and DataFrame columns
- New NaT (Not-a-Time) type to represent NA in timestamp arrays
- Optimize Series.asof for looking up “as of” values for arrays of timestamps
- Milli, Micro, Nano date offset objects
- Can index time series with datetime.time objects to select all data at
particular time of day (
TimeSeries.at_time) or between two times (TimeSeries.between_time) - Add tshift method for leading/lagging using the frequency (if any) of the index, as opposed to a naive lead/lag using shift
Other new features¶
- New cut and
qcutfunctions (like R’s cut function) for computing a categorical variable from a continuous variable by binning values either into value-based (cut) or quantile-based (qcut) bins - Rename
FactortoCategoricaland add a number of usability features - Add limit argument to fillna/reindex
- More flexible multiple function application in GroupBy, and can pass list (name, function) tuples to get result in particular order with given names
- Add flexible replace method for efficiently substituting values
- Enhanced read_csv/read_table for reading time series data and converting multiple columns to dates
- Add comments option to parser functions: read_csv, etc.
- Add :ref`dayfirst <io.dayfirst>` option to parser functions for parsing international DD/MM/YYYY dates
- Allow the user to specify the CSV reader dialect to control quoting etc.
- Handling thousands separators in read_csv to improve integer parsing.
- Enable unstacking of multiple levels in one shot. Alleviate
pivot_tablebugs (empty columns being introduced) - Move to klib-based hash tables for indexing; better performance and less memory usage than Python’s dict
- Add first, last, min, max, and prod optimized GroupBy functions
- New ordered_merge function
- Add flexible comparison instance methods eq, ne, lt, gt, etc. to DataFrame, Series
- Improve scatter_matrix plotting function and add histogram or kernel density estimates to diagonal
- Add ‘kde’ plot option for density plots
- Support for converting DataFrame to R data.frame through rpy2
- Improved support for complex numbers in Series and DataFrame
- Add pct_change method to all data structures
- Add max_colwidth configuration option for DataFrame console output
- Interpolate Series values using index values
- Can select multiple columns from GroupBy
- Add update methods to Series/DataFrame for updating values in place
- Add
anyandallmethod to DataFrame
New plotting methods¶
Series.plot now supports a secondary_y option:
In [1]: plt.figure()
Out[1]: <matplotlib.figure.Figure at 0x133e4b8d0>
In [2]: fx['FR'].plot(style='g')
��������������������������������������������������Out[2]: <matplotlib.axes._subplots.AxesSubplot at 0x12ad13cc0>
In [3]: fx['IT'].plot(style='k--', secondary_y=True)
�����������������������������������������������������������������������������������������������������������������Out[3]: <matplotlib.axes._subplots.AxesSubplot at 0x133554dd8>
Vytautas Jancauskas, the 2012 GSOC participant, has added many new plot
types. For example, 'kde' is a new option:
In [4]: s = Series(np.concatenate((np.random.randn(1000),
...: np.random.randn(1000) * 0.5 + 3)))
...:
In [5]: plt.figure()
Out[5]: <matplotlib.figure.Figure at 0x1300adda0>
In [6]: s.hist(normed=True, alpha=0.2)
��������������������������������������������������Out[6]: <matplotlib.axes._subplots.AxesSubplot at 0x13099ca20>
In [7]: s.plot(kind='kde')
�����������������������������������������������������������������������������������������������������������������Out[7]: <matplotlib.axes._subplots.AxesSubplot at 0x13099ca20>
See the plotting page for much more.
Other API changes¶
- Deprecation of
offset,time_rule, andtimeRulearguments names in time series functions. Warnings will be printed until pandas 0.9 or 1.0.
Potential porting issues for pandas <= 0.7.3 users¶
The major change that may affect you in pandas 0.8.0 is that time series
indexes use NumPy’s datetime64 data type instead of dtype=object arrays
of Python’s built-in datetime.datetime objects. DateRange has been
replaced by DatetimeIndex but otherwise behaved identically. But, if you
have code that converts DateRange or Index objects that used to contain
datetime.datetime values to plain NumPy arrays, you may have bugs lurking
with code using scalar values because you are handing control over to NumPy:
In [8]: import datetime
In [9]: rng = date_range('1/1/2000', periods=10)
In [10]: rng[5]
Out[10]: Timestamp('2000-01-06 00:00:00', freq='D')
In [11]: isinstance(rng[5], datetime.datetime)
����������������������������������������������������Out[11]: True
In [12]: rng_asarray = np.asarray(rng)
In [13]: scalar_val = rng_asarray[5]
In [14]: type(scalar_val)
Out[14]: numpy.datetime64
pandas’s Timestamp object is a subclass of datetime.datetime that has
nanosecond support (the nanosecond field store the nanosecond value between
0 and 999). It should substitute directly into any code that used
datetime.datetime values before. Thus, I recommend not casting
DatetimeIndex to regular NumPy arrays.
If you have code that requires an array of datetime.datetime objects, you
have a couple of options. First, the asobject property of DatetimeIndex
produces an array of Timestamp objects:
In [15]: stamp_array = rng.asobject
In [16]: stamp_array
Out[16]:
Index([2000-01-01 00:00:00, 2000-01-02 00:00:00, 2000-01-03 00:00:00,
2000-01-04 00:00:00, 2000-01-05 00:00:00, 2000-01-06 00:00:00,
2000-01-07 00:00:00, 2000-01-08 00:00:00, 2000-01-09 00:00:00,
2000-01-10 00:00:00],
dtype='object')
In [17]: stamp_array[5]
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[17]: Timestamp('2000-01-06 00:00:00', freq='D')
To get an array of proper datetime.datetime objects, use the
to_pydatetime method:
In [18]: dt_array = rng.to_pydatetime()
In [19]: dt_array
Out[19]:
array([datetime.datetime(2000, 1, 1, 0, 0),
datetime.datetime(2000, 1, 2, 0, 0),
datetime.datetime(2000, 1, 3, 0, 0),
datetime.datetime(2000, 1, 4, 0, 0),
datetime.datetime(2000, 1, 5, 0, 0),
datetime.datetime(2000, 1, 6, 0, 0),
datetime.datetime(2000, 1, 7, 0, 0),
datetime.datetime(2000, 1, 8, 0, 0),
datetime.datetime(2000, 1, 9, 0, 0),
datetime.datetime(2000, 1, 10, 0, 0)], dtype=object)
In [20]: dt_array[5]
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[20]: datetime.datetime(2000, 1, 6, 0, 0)
matplotlib knows how to handle datetime.datetime but not Timestamp
objects. While I recommend that you plot time series using TimeSeries.plot,
you can either use to_pydatetime or register a converter for the Timestamp
type. See matplotlib documentation for more on this.
Warning
There are bugs in the user-facing API with the nanosecond datetime64 unit
in NumPy 1.6. In particular, the string version of the array shows garbage
values, and conversion to dtype=object is similarly broken.
In [21]: rng = date_range('1/1/2000', periods=10)
In [22]: rng
Out[22]:
DatetimeIndex(['2000-01-01', '2000-01-02', '2000-01-03', '2000-01-04',
'2000-01-05', '2000-01-06', '2000-01-07', '2000-01-08',
'2000-01-09', '2000-01-10'],
dtype='datetime64[ns]', freq='D')
In [23]: np.asarray(rng)
����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[23]:
array(['2000-01-01T00:00:00.000000000', '2000-01-02T00:00:00.000000000',
'2000-01-03T00:00:00.000000000', '2000-01-04T00:00:00.000000000',
'2000-01-05T00:00:00.000000000', '2000-01-06T00:00:00.000000000',
'2000-01-07T00:00:00.000000000', '2000-01-08T00:00:00.000000000',
'2000-01-09T00:00:00.000000000', '2000-01-10T00:00:00.000000000'], dtype='datetime64[ns]')
In [24]: converted = np.asarray(rng, dtype=object)
In [25]: converted[5]
Out[25]: 947116800000000000
Trust me: don’t panic. If you are using NumPy 1.6 and restrict your
interaction with datetime64 values to pandas’s API you will be just
fine. There is nothing wrong with the data-type (a 64-bit integer
internally); all of the important data processing happens in pandas and is
heavily tested. I strongly recommend that you do not work directly with
datetime64 arrays in NumPy 1.6 and only use the pandas API.
Support for non-unique indexes: In the latter case, you may have code
inside a try:... catch: block that failed due to the index not being
unique. In many cases it will no longer fail (some method like append still
check for uniqueness unless disabled). However, all is not lost: you can
inspect index.is_unique and raise an exception explicitly if it is
False or go to a different code branch.
v.0.7.3 (April 12, 2012)¶
This is a minor release from 0.7.2 and fixes many minor bugs and adds a number of nice new features. There are also a couple of API changes to note; these should not affect very many users, and we are inclined to call them “bug fixes” even though they do constitute a change in behavior. See the full release notes or issue tracker on GitHub for a complete list.
New features¶
- New fixed width file reader,
read_fwf - New scatter_matrix function for making a scatter plot matrix
from pandas.tools.plotting import scatter_matrix
scatter_matrix(df, alpha=0.2)
- Add
stackedargument to Series and DataFrame’splotmethod for stacked bar plots.
df.plot(kind='bar', stacked=True)
df.plot(kind='barh', stacked=True)
- Add log x and y scaling options to
DataFrame.plotandSeries.plot - Add
kurtmethods to Series and DataFrame for computing kurtosis
NA Boolean Comparison API Change¶
Reverted some changes to how NA values (represented typically as NaN or
None) are handled in non-numeric Series:
In [1]: series = Series(['Steve', np.nan, 'Joe'])
In [2]: series == 'Steve'
Out[2]:
0 True
1 False
2 False
Length: 3, dtype: bool
In [3]: series != 'Steve'
�����������������������������������������������������������������Out[3]:
0 False
1 True
2 True
Length: 3, dtype: bool
In comparisons, NA / NaN will always come through as False except with
!= which is True. Be very careful with boolean arithmetic, especially
negation, in the presence of NA data. You may wish to add an explicit NA
filter into boolean array operations if you are worried about this:
In [4]: mask = series == 'Steve'
In [5]: series[mask & series.notnull()]
Out[5]:
0 Steve
Length: 1, dtype: object
While propagating NA in comparisons may seem like the right behavior to some users (and you could argue on purely technical grounds that this is the right thing to do), the evaluation was made that propagating NA everywhere, including in numerical arrays, would cause a large amount of problems for users. Thus, a “practicality beats purity” approach was taken. This issue may be revisited at some point in the future.
Other API Changes¶
When calling apply on a grouped Series, the return value will also be a
Series, to be more consistent with the groupby behavior with DataFrame:
In [6]: df = DataFrame({'A' : ['foo', 'bar', 'foo', 'bar',
...: 'foo', 'bar', 'foo', 'foo'],
...: 'B' : ['one', 'one', 'two', 'three',
...: 'two', 'two', 'one', 'three'],
...: 'C' : np.random.randn(8), 'D' : np.random.randn(8)})
...:
In [7]: df
Out[7]:
A B C D
0 foo one 1.075059 -0.449141
1 bar one 0.785676 1.443014
2 foo two 0.958157 0.612324
3 bar three 1.477773 -0.178818
4 foo two -1.006023 0.133072
5 bar two -1.506997 -0.550981
6 foo one 1.218042 -2.043335
7 foo three -0.565878 0.753539
[8 rows x 4 columns]
In [8]: grouped = df.groupby('A')['C']
In [9]: grouped.describe()
Out[9]:
count mean std min 25% 50% 75% \
A
bar 3.0 0.252151 1.562274 -1.506997 -0.360661 0.785676 1.131724
foo 5.0 0.335871 1.039915 -1.006023 -0.565878 0.958157 1.075059
max
A
bar 1.477773
foo 1.218042
[2 rows x 8 columns]
In [10]: grouped.apply(lambda x: x.sort_values()[-2:]) # top 2 values
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[10]:
A
bar 1 0.785676
3 1.477773
foo 0 1.075059
6 1.218042
Name: C, Length: 4, dtype: float64
v.0.7.2 (March 16, 2012)¶
This release targets bugs in 0.7.1, and adds a few minor features.
New features¶
- Add additional tie-breaking methods in DataFrame.rank (GH874)
- Add ascending parameter to rank in Series, DataFrame (GH875)
- Add coerce_float option to DataFrame.from_records (GH893)
- Add sort_columns parameter to allow unsorted plots (GH918)
- Enable column access via attributes on GroupBy (GH882)
- Can pass dict of values to DataFrame.fillna (GH661)
- Can select multiple hierarchical groups by passing list of values in .ix (GH134)
- Add
axisoption to DataFrame.fillna (GH174)- Add level keyword to
dropfor dropping values from a level (GH159)
v.0.7.1 (February 29, 2012)¶
This release includes a few new features and addresses over a dozen bugs in 0.7.0.
New features¶
- Add
to_clipboardfunction to pandas namespace for writing objects to the system clipboard (GH774)- Add
itertuplesmethod to DataFrame for iterating through the rows of a dataframe as tuples (GH818)- Add ability to pass fill_value and method to DataFrame and Series align method (GH806, GH807)
- Add fill_value option to reindex, align methods (GH784)
- Enable concat to produce DataFrame from Series (GH787)
- Add
betweenmethod to Series (GH802)- Add HTML representation hook to DataFrame for the IPython HTML notebook (GH773)
- Support for reading Excel 2007 XML documents using openpyxl
v.0.7.0 (February 9, 2012)¶
New features¶
- New unified merge function for efficiently performing full gamut of database / relational-algebra operations. Refactored existing join methods to use the new infrastructure, resulting in substantial performance gains (GH220, GH249, GH267)
- New unified concatenation function for concatenating
Series, DataFrame or Panel objects along an axis. Can form union or
intersection of the other axes. Improves performance of
Series.appendandDataFrame.append(GH468, GH479, GH273) - Can pass multiple DataFrames to
DataFrame.append to concatenate (stack) and multiple Series to
Series.appendtoo - Can pass list of dicts (e.g., a list of JSON objects) to DataFrame constructor (GH526)
- You can now set multiple columns in a
DataFrame via
__getitem__, useful for transformation (GH342) - Handle differently-indexed output values in
DataFrame.apply(GH498)
In [1]: df = DataFrame(randn(10, 4))
In [2]: df.apply(lambda x: x.describe())
Out[2]:
0 1 2 3
count 10.000000 10.000000 10.000000 10.000000
mean -0.409608 0.539495 0.163276 0.051646
std 1.397779 0.968808 0.874489 0.719651
min -2.539411 -0.737206 -1.202276 -1.050435
25% -1.202202 0.021308 -0.368812 -0.383608
50% -0.384480 0.306124 0.211431 0.165586
75% 0.186280 1.024039 0.730744 0.494457
max 2.524998 2.533114 1.334428 1.147396
[8 rows x 4 columns]
- Add
reorder_levelsmethod to Series and DataFrame (GH534) - Add dict-like
getfunction to DataFrame and Panel (GH521) - Add
DataFrame.iterrowsmethod for efficiently iterating through the rows of a DataFrame - Add
DataFrame.to_panelwith code adapted fromLongPanel.to_long - Add
reindex_axismethod added to DataFrame - Add
leveloption to binary arithmetic functions onDataFrameandSeries - Add
leveloption to thereindexandalignmethods on Series and DataFrame for broadcasting values across a level (GH542, GH552, others) - Add attribute-based item access to
Paneland add IPython completion (GH563) - Add
logyoption toSeries.plotfor log-scaling on the Y axis - Add
indexandheaderoptions toDataFrame.to_string - Can pass multiple DataFrames to
DataFrame.jointo join on index (GH115) - Can pass multiple Panels to
Panel.join(GH115) - Added
justifyargument toDataFrame.to_stringto allow different alignment of column headers - Add
sortoption to GroupBy to allow disabling sorting of the group keys for potential speedups (GH595) - Can pass MaskedArray to Series constructor (GH563)
- Add Panel item access via attributes and IPython completion (GH554)
- Implement
DataFrame.lookup, fancy-indexing analogue for retrieving values given a sequence of row and column labels (GH338) - Can pass a list of functions to aggregate with groupby on a DataFrame, yielding an aggregated result with hierarchical columns (GH166)
- Can call
cumminandcummaxon Series and DataFrame to get cumulative minimum and maximum, respectively (GH647) value_rangeadded as utility function to get min and max of a dataframe (GH288)- Added
encodingargument toread_csv,read_table,to_csvandfrom_csvfor non-ascii text (GH717) - Added
absmethod to pandas objects - Added
crosstabfunction for easily computing frequency tables - Added
isinmethod to index objects - Added
levelargument toxsmethod of DataFrame.
API Changes to integer indexing¶
One of the potentially riskiest API changes in 0.7.0, but also one of the most important, was a complete review of how integer indexes are handled with regard to label-based indexing. Here is an example:
In [3]: s = Series(randn(10), index=range(0, 20, 2))
In [4]: s
Out[4]:
0 -0.543429
2 1.425447
4 -0.408795
6 -1.489348
8 -1.166408
10 -0.481205
12 -0.810355
14 -0.985491
16 -0.336246
18 -0.629058
Length: 10, dtype: float64
In [5]: s[0]
������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[5]: -0.54342898765020686
In [6]: s[2]
�����������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[6]: 1.4254474252163707
In [7]: s[4]
��������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[7]: -0.40879476802408349
This is all exactly identical to the behavior before. However, if you ask for a
key not contained in the Series, in versions 0.6.1 and prior, Series would
fall back on a location-based lookup. This now raises a KeyError:
In [2]: s[1]
KeyError: 1
This change also has the same impact on DataFrame:
In [3]: df = DataFrame(randn(8, 4), index=range(0, 16, 2))
In [4]: df
0 1 2 3
0 0.88427 0.3363 -0.1787 0.03162
2 0.14451 -0.1415 0.2504 0.58374
4 -1.44779 -0.9186 -1.4996 0.27163
6 -0.26598 -2.4184 -0.2658 0.11503
8 -0.58776 0.3144 -0.8566 0.61941
10 0.10940 -0.7175 -1.0108 0.47990
12 -1.16919 -0.3087 -0.6049 -0.43544
14 -0.07337 0.3410 0.0424 -0.16037
In [5]: df.ix[3]
KeyError: 3
In order to support purely integer-based indexing, the following methods have been added:
| Method | Description |
|---|---|
Series.iget_value(i) |
Retrieve value stored at location i |
Series.iget(i) |
Alias for iget_value |
DataFrame.irow(i) |
Retrieve the i-th row |
DataFrame.icol(j) |
Retrieve the j-th column |
DataFrame.iget_value(i, j) |
Retrieve the value at row i and column j |
API tweaks regarding label-based slicing¶
Label-based slicing using ix now requires that the index be sorted
(monotonic) unless both the start and endpoint are contained in the index:
In [1]: s = Series(randn(6), index=list('gmkaec'))
In [2]: s
Out[2]:
g -1.182230
m -0.276183
k -0.243550
a 1.628992
e 0.073308
c -0.539890
dtype: float64
Then this is OK:
In [3]: s.ix['k':'e']
Out[3]:
k -0.243550
a 1.628992
e 0.073308
dtype: float64
But this is not:
In [12]: s.ix['b':'h']
KeyError 'b'
If the index had been sorted, the “range selection” would have been possible:
In [4]: s2 = s.sort_index()
In [5]: s2
Out[5]:
a 1.628992
c -0.539890
e 0.073308
g -1.182230
k -0.243550
m -0.276183
dtype: float64
In [6]: s2.ix['b':'h']
Out[6]:
c -0.539890
e 0.073308
g -1.182230
dtype: float64
Changes to Series [] operator¶
As as notational convenience, you can pass a sequence of labels or a label
slice to a Series when getting and setting values via [] (i.e. the
__getitem__ and __setitem__ methods). The behavior will be the same as
passing similar input to ix except in the case of integer indexing:
In [8]: s = Series(randn(6), index=list('acegkm'))
In [9]: s
Out[9]:
a -0.297788
c 0.499769
e 0.810531
g 0.414649
k -1.551478
m 1.012459
Length: 6, dtype: float64
In [10]: s[['m', 'a', 'c', 'e']]
�����������������������������������������������������������������������������������������������������������������������Out[10]:
m 1.012459
a -0.297788
c 0.499769
e 0.810531
Length: 4, dtype: float64
In [11]: s['b':'l']
�������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[11]:
c 0.499769
e 0.810531
g 0.414649
k -1.551478
Length: 4, dtype: float64
In [12]: s['c':'k']
���������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������������Out[12]:
c 0.499769
e 0.810531
g 0.414649
k -1.551478
Length: 4, dtype: float64
In the case of integer indexes, the behavior will be exactly as before
(shadowing ndarray):
In [13]: s = Series(randn(6), index=range(0, 12, 2))
In [14]: s[[4, 0, 2]]
Out[14]:
4 0.928877
0 1.171752
2 0.026488
Length: 3, dtype: float64
In [15]: s[1:5]
������������������������������������������������������������������������������Out[15]:
2 0.026488
4 0.928877
6 -1.264991
8 0.419449
Length: 4, dtype: float64
If you wish to do indexing with sequences and slicing on an integer index with
label semantics, use ix.
Other API Changes¶
- The deprecated
LongPanelclass has been completely removed - If
Series.sortis called on a column of a DataFrame, an exception will now be raised. Before it was possible to accidentally mutate a DataFrame’s column by doingdf[col].sort()instead of the side-effect free methoddf[col].order()(GH316) - Miscellaneous renames and deprecations which will (harmlessly) raise
FutureWarning dropadded as an optional parameter toDataFrame.reset_index(GH699)
Performance improvements¶
- Cythonized GroupBy aggregations no longer presort the data, thus achieving a significant speedup (GH93). GroupBy aggregations with Python functions significantly sped up by clever manipulation of the ndarray data type in Cython (GH496).
- Better error message in DataFrame constructor when passed column labels don’t match data (GH497)
- Substantially improve performance of multi-GroupBy aggregation when a Python function is passed, reuse ndarray object in Cython (GH496)
- Can store objects indexed by tuples and floats in HDFStore (GH492)
- Don’t print length by default in Series.to_string, add length option (GH489)
- Improve Cython code for multi-groupby to aggregate without having to sort the data (GH93)
- Improve MultiIndex reindexing speed by storing tuples in the MultiIndex, test for backwards unpickling compatibility
- Improve column reindexing performance by using specialized Cython take function
- Further performance tweaking of Series.__getitem__ for standard use cases
- Avoid Index dict creation in some cases (i.e. when getting slices, etc.), regression from prior versions
- Friendlier error message in setup.py if NumPy not installed
- Use common set of NA-handling operations (sum, mean, etc.) in Panel class also (GH536)
- Default name assignment when calling
reset_indexon DataFrame with a regular (non-hierarchical) index (GH476) - Use Cythonized groupers when possible in Series/DataFrame stat ops with
levelparameter passed (GH545) - Ported skiplist data structure to C to speed up
rolling_medianby about 5-10x in most typical use cases (GH374)
v.0.6.1 (December 13, 2011)¶
New features¶
- Can append single rows (as Series) to a DataFrame
- Add Spearman and Kendall rank correlation options to Series.corr and DataFrame.corr (GH428)
- Added
get_valueandset_valuemethods to Series, DataFrame, and Panel for very low-overhead access (>2x faster in many cases) to scalar elements (GH437, GH438).set_valueis capable of producing an enlarged object. - Add PyQt table widget to sandbox (GH435)
- DataFrame.align can accept Series arguments and an axis option (GH461)
- Implement new SparseArray and SparseList data structures. SparseSeries now derives from SparseArray (GH463)
- Better console printing options (GH453)
- Implement fast data ranking for Series and DataFrame, fast versions of scipy.stats.rankdata (GH428)
- Implement DataFrame.from_items alternate constructor (GH444)
- DataFrame.convert_objects method for inferring better dtypes for object columns (GH302)
- Add rolling_corr_pairwise function for computing Panel of correlation matrices (GH189)
- Add margins option to pivot_table for computing subgroup aggregates (GH114)
- Add
Series.from_csvfunction (GH482) - Can pass DataFrame/DataFrame and DataFrame/Series to rolling_corr/rolling_cov (GH #462)
- MultiIndex.get_level_values can accept the level name
Performance improvements¶
- Improve memory usage of DataFrame.describe (do not copy data unnecessarily) (PR #425)
- Optimize scalar value lookups in the general case by 25% or more in Series and DataFrame
- Fix performance regression in cross-sectional count in DataFrame, affecting DataFrame.dropna speed
- Column deletion in DataFrame copies no data (computes views on blocks) (GH #158)
v.0.6.0 (November 25, 2011)¶
New Features¶
- Added
meltfunction topandas.core.reshape - Added
levelparameter to group by level in Series and DataFrame descriptive statistics (GH313) - Added
headandtailmethods to Series, analogous to to DataFrame (GH296) - Added
Series.isinfunction which checks if each value is contained in a passed sequence (GH289) - Added
float_formatoption toSeries.to_string - Added
skip_footer(GH291) andconverters(GH343) options toread_csvandread_table - Added
drop_duplicatesandduplicatedfunctions for removing duplicate DataFrame rows and checking for duplicate rows, respectively (GH319) - Implemented operators ‘&’, ‘|’, ‘^’, ‘-‘ on DataFrame (GH347)
- Added
Series.mad, mean absolute deviation - Added
QuarterEndDateOffset (GH321) - Added
dotto DataFrame (GH65) - Added
orientoption toPanel.from_dict(GH359, GH301) - Added
orientoption toDataFrame.from_dict - Added passing list of tuples or list of lists to
DataFrame.from_records(GH357) - Added multiple levels to groupby (GH103)
- Allow multiple columns in
byargument ofDataFrame.sort_index(GH92, GH362) - Added fast
get_valueandput_valuemethods to DataFrame (GH360) - Added
covinstance methods to Series and DataFrame (GH194, GH362) - Added
kind='bar'option toDataFrame.plot(GH348) - Added
idxminandidxmaxto Series and DataFrame (GH286) - Added
read_clipboardfunction to parse DataFrame from clipboard (GH300) - Added
nuniquefunction to Series for counting unique elements (GH297) - Made DataFrame constructor use Series name if no columns passed (GH373)
- Support regular expressions in read_table/read_csv (GH364)
- Added
DataFrame.to_htmlfor writing DataFrame to HTML (GH387) - Added support for MaskedArray data in DataFrame, masked values converted to NaN (GH396)
- Added
DataFrame.boxplotfunction (GH368) - Can pass extra args, kwds to DataFrame.apply (GH376)
- Implement
DataFrame.joinwith vectoronargument (GH312) - Added
legendboolean flag toDataFrame.plot(GH324) - Can pass multiple levels to
stackandunstack(GH370) - Can pass multiple values columns to
pivot_table(GH381) - Use Series name in GroupBy for result index (GH363)
- Added
rawoption toDataFrame.applyfor performance if only need ndarray (GH309) - Added proper, tested weighted least squares to standard and panel OLS (GH303)
Performance Enhancements¶
- VBENCH Cythonized
cache_readonly, resulting in substantial micro-performance enhancements throughout the codebase (GH361) - VBENCH Special Cython matrix iterator for applying arbitrary reduction operations with 3-5x better performance than np.apply_along_axis (GH309)
- VBENCH Improved performance of
MultiIndex.from_tuples - VBENCH Special Cython matrix iterator for applying arbitrary reduction operations
- VBENCH + DOCUMENT Add
rawoption toDataFrame.applyfor getting better performance when - VBENCH Faster cythonized count by level in Series and DataFrame (GH341)
- VBENCH? Significant GroupBy performance enhancement with multiple keys with many “empty” combinations
- VBENCH New Cython vectorized function
map_inferspeeds upSeries.applyandSeries.mapsignificantly when passed elementwise Python function, motivated by (GH355) - VBENCH Significantly improved performance of
Series.order, which also makes np.unique called on a Series faster (GH327) - VBENCH Vastly improved performance of GroupBy on axes with a MultiIndex (GH299)
v.0.5.0 (October 24, 2011)¶
New Features¶
- Added
DataFrame.alignmethod with standard join options - Added
parse_datesoption toread_csvandread_tablemethods to optionally try to parse dates in the index columns - Added
nrows,chunksize, anditeratorarguments toread_csvandread_table. The last two return a newTextParserclass capable of lazily iterating through chunks of a flat file (GH242) - Added ability to join on multiple columns in
DataFrame.join(GH214) - Added private
_get_duplicatesfunction toIndexfor identifying duplicate values more easily (ENH5c) - Added column attribute access to DataFrame.
- Added Python tab completion hook for DataFrame columns. (GH233, GH230)
- Implemented
Series.describefor Series containing objects (GH241) - Added inner join option to
DataFrame.joinwhen joining on key(s) (GH248) - Implemented selecting DataFrame columns by passing a list to
__getitem__(GH253) - Implemented & and | to intersect / union Index objects, respectively (GH261)
- Added
pivot_tableconvenience function to pandas namespace (GH234) - Implemented
Panel.rename_axisfunction (GH243) - DataFrame will show index level names in console output (GH334)
- Implemented
Panel.take - Added
set_eng_float_formatfor alternate DataFrame floating point string formatting (ENH61) - Added convenience
set_indexfunction for creating a DataFrame index from its existing columns - Implemented
groupbyhierarchical index level name (GH223) - Added support for different delimiters in
DataFrame.to_csv(GH244) - TODO: DOCS ABOUT TAKE METHODS
Performance Enhancements¶
- VBENCH Major performance improvements in file parsing functions
read_csvandread_table - VBENCH Added Cython function for converting tuples to ndarray very fast. Speeds up many MultiIndex-related operations
- VBENCH Refactored merging / joining code into a tidy class and disabled unnecessary computations in the float/object case, thus getting about 10% better performance (GH211)
- VBENCH Improved speed of
DataFrame.xson mixed-type DataFrame objects by about 5x, regression from 0.3.0 (GH215) - VBENCH With new
DataFrame.alignmethod, speeding up binary operations between differently-indexed DataFrame objects by 10-25%. - VBENCH Significantly sped up conversion of nested dict into DataFrame (GH212)
- VBENCH Significantly speed up DataFrame
__repr__andcounton large mixed-type DataFrame objects
v.0.4.3 through v0.4.1 (September 25 - October 9, 2011)¶
New Features¶
- Added Python 3 support using 2to3 (GH200)
- Added
nameattribute toSeries, now prints as part ofSeries.__repr__ - Added instance methods
isnullandnotnullto Series (GH209, GH203) - Added
Series.alignmethod for aligning two series with choice of join method (ENH56) - Added method
get_level_valuestoMultiIndex(GH188) - Set values in mixed-type
DataFrameobjects via.ixindexing attribute (GH135) - Added new
DataFramemethodsget_dtype_countsand propertydtypes(ENHdc) - Added ignore_index option to
DataFrame.appendto stack DataFrames (ENH1b) read_csvtries to sniff delimiters usingcsv.Sniffer(GH146)read_csvcan read multiple columns into aMultiIndex; DataFrame’sto_csvmethod writes out a correspondingMultiIndex(GH151)DataFrame.renamehas a newcopyparameter to rename a DataFrame in place (ENHed)- Enable unstacking by name (GH142)
- Enable
sortlevelto work by level (GH141)
Performance Enhancements¶
- Altered binary operations on differently-indexed SparseSeries objects to use the integer-based (dense) alignment logic which is faster with a larger number of blocks (GH205)
- Wrote faster Cython data alignment / merging routines resulting in substantial speed increases
- Improved performance of
isnullandnotnull, a regression from v0.3.0 (GH187) - Refactored code related to
DataFrame.joinso that intermediate aligned copies of the data in eachDataFrameargument do not need to be created. Substantial performance increases result (GH176) - Substantially improved performance of generic
Index.intersectionandIndex.union - Implemented
BlockManager.takeresulting in significantly fastertakeperformance on mixed-typeDataFrameobjects (GH104) - Improved performance of
Series.sort_index - Significant groupby performance enhancement: removed unnecessary integrity checks in DataFrame internals that were slowing down slicing operations to retrieve groups
- Optimized
_ensure_indexfunction resulting in performance savings in type-checking Index objects - Wrote fast time series merging / joining methods in Cython. Will be integrated later into DataFrame.join and related functions