Working with missing data¶
In this section, we will discuss missing (also referred to as NA) values in pandas.
Note
The choice of using NaN internally to denote missing data was largely for simplicity and performance reasons. It differs from the MaskedArray approach of, for example, scikits.timeseries. We are hopeful that NumPy will soon be able to provide a native NA type solution (similar to R) performant enough to be used in pandas.
Missing data basics¶
When / why does data become missing?¶
Some might quibble over our usage of missing. By “missing” we simply mean null or “not present for whatever reason”. Many data sets simply arrive with missing data, either because it exists and was not collected or it never existed. For example, in a collection of financial time series, some of the time series might start on different dates. Thus, values prior to the start date would generally be marked as missing.
In pandas, one of the most common ways that missing data is introduced into a data set is by reindexing. For example
In [925]: df = DataFrame(randn(5, 3), index=['a', 'c', 'e', 'f', 'h'],
.....: columns=['one', 'two', 'three'])
.....:
In [926]: df['four'] = 'bar'
In [927]: df['five'] = df['one'] > 0
In [928]: df
Out[928]:
one two three four five
a 0.059117 1.138469 -2.400634 bar True
c -0.280853 0.025653 -1.386071 bar False
e 0.863937 0.252462 1.500571 bar True
f 1.053202 -2.338595 -0.374279 bar True
h -2.359958 -1.157886 -0.551865 bar False
In [929]: df2 = df.reindex(['a', 'b', 'c', 'd', 'e', 'f', 'g', 'h'])
In [930]: df2
Out[930]:
one two three four five
a 0.059117 1.138469 -2.400634 bar True
b NaN NaN NaN NaN NaN
c -0.280853 0.025653 -1.386071 bar False
d NaN NaN NaN NaN NaN
e 0.863937 0.252462 1.500571 bar True
f 1.053202 -2.338595 -0.374279 bar True
g NaN NaN NaN NaN NaN
h -2.359958 -1.157886 -0.551865 bar False
Values considered “missing”¶
As data comes in many shapes and forms, pandas aims to be flexible with regard to handling missing data. While NaN is the default missing value marker for reasons of computational speed and convenience, we need to be able to easily detect this value with data of different types: floating point, integer, boolean, and general object. In many cases, however, the Python None will arise and we wish to also consider that “missing” or “null”. Lastly, for legacy reasons inf and -inf are also considered to be “null” in computations. Since in NumPy divide-by-zero generates inf or -inf and not NaN, I think you will find this is a worthwhile trade-off (Zen of Python: “practicality beats purity”).
To make detecting missing values easier (and across different array dtypes), pandas provides the isnull() and notnull() functions, which are also methods on Series objects:
In [931]: df2['one']
Out[931]:
a 0.059117
b NaN
c -0.280853
d NaN
e 0.863937
f 1.053202
g NaN
h -2.359958
Name: one
In [932]: isnull(df2['one'])
Out[932]:
a False
b True
c False
d True
e False
f False
g True
h False
Name: one
In [933]: df2['four'].notnull()
Out[933]:
a True
b False
c True
d False
e True
f True
g False
h True
Summary: NaN, inf, -inf, and None (in object arrays) are all considered missing by the isnull and notnull functions.
Calculations with missing data¶
Missing values propagate naturally through arithmetic operations between pandas objects.
In [934]: a
Out[934]:
one two
a 0.059117 1.138469
b 0.059117 1.138469
c -0.280853 0.025653
d -0.280853 0.025653
e 0.863937 0.252462
In [935]: b
Out[935]:
one two three
a 0.059117 1.138469 -2.400634
b NaN NaN NaN
c -0.280853 0.025653 -1.386071
d NaN NaN NaN
e 0.863937 0.252462 1.500571
In [936]: a + b
Out[936]:
one three two
a 0.118234 NaN 2.276938
b NaN NaN NaN
c -0.561707 NaN 0.051306
d NaN NaN NaN
e 1.727874 NaN 0.504923
The descriptive statistics and computational methods discussed in the data structure overview (and listed here and here) are all written to account for missing data. For example:
- When summing data, NA (missing) values will be treated as zero
- If the data are all NA, the result will be NA
- Methods like cumsum and cumprod ignore NA values, but preserve them in the resulting arrays
In [937]: df
Out[937]:
one two three
a 0.059117 1.138469 -2.400634
b NaN NaN NaN
c -0.280853 0.025653 -1.386071
d NaN NaN NaN
e 0.863937 0.252462 1.500571
f 1.053202 -2.338595 -0.374279
g NaN NaN NaN
h -2.359958 -1.157886 -0.551865
In [938]: df['one'].sum()
Out[938]: -0.66455558290247652
In [939]: df.mean(1)
Out[939]:
a -0.401016
b NaN
c -0.547090
d NaN
e 0.872323
f -0.553224
g NaN
h -1.356570
In [940]: df.cumsum()
Out[940]:
one two three
a 0.059117 1.138469 -2.400634
b NaN NaN NaN
c -0.221736 1.164122 -3.786705
d NaN NaN NaN
e 0.642200 1.416584 -2.286134
f 1.695403 -0.922011 -2.660413
g NaN NaN NaN
h -0.664556 -2.079897 -3.212278
NA values in GroupBy¶
NA groups in GroupBy are automatically excluded. This behavior is consistent with R, for example.
Cleaning / filling missing data¶
pandas objects are equipped with various data manipulation methods for dealing with missing data.
Filling missing values: fillna¶
The fillna function can “fill in” NA values with non-null data in a couple of ways, which we illustrate:
Replace NA with a scalar value
In [941]: df2
Out[941]:
one two three four five
a 0.059117 1.138469 -2.400634 bar True
b NaN NaN NaN NaN NaN
c -0.280853 0.025653 -1.386071 bar False
d NaN NaN NaN NaN NaN
e 0.863937 0.252462 1.500571 bar True
f 1.053202 -2.338595 -0.374279 bar True
g NaN NaN NaN NaN NaN
h -2.359958 -1.157886 -0.551865 bar False
In [942]: df2.fillna(0)
Out[942]:
one two three four five
a 0.059117 1.138469 -2.400634 bar True
b 0.000000 0.000000 0.000000 0 0
c -0.280853 0.025653 -1.386071 bar False
d 0.000000 0.000000 0.000000 0 0
e 0.863937 0.252462 1.500571 bar True
f 1.053202 -2.338595 -0.374279 bar True
g 0.000000 0.000000 0.000000 0 0
h -2.359958 -1.157886 -0.551865 bar False
In [943]: df2['four'].fillna('missing')
Out[943]:
a bar
b missing
c bar
d missing
e bar
f bar
g missing
h bar
Name: four
Fill gaps forward or backward
Using the same filling arguments as reindexing, we can propagate non-null values forward or backward:
In [944]: df
Out[944]:
one two three
a 0.059117 1.138469 -2.400634
b NaN NaN NaN
c -0.280853 0.025653 -1.386071
d NaN NaN NaN
e 0.863937 0.252462 1.500571
f 1.053202 -2.338595 -0.374279
g NaN NaN NaN
h -2.359958 -1.157886 -0.551865
In [945]: df.fillna(method='pad')
Out[945]:
one two three
a 0.059117 1.138469 -2.400634
b 0.059117 1.138469 -2.400634
c -0.280853 0.025653 -1.386071
d -0.280853 0.025653 -1.386071
e 0.863937 0.252462 1.500571
f 1.053202 -2.338595 -0.374279
g 1.053202 -2.338595 -0.374279
h -2.359958 -1.157886 -0.551865
Limit the amount of filling
If we only want consecutive gaps filled up to a certain number of data points, we can use the limit keyword:
In [946]: df
Out[946]:
one two three
a 0.059117 1.138469 -2.400634
b NaN NaN NaN
c NaN NaN NaN
d NaN NaN NaN
e 0.863937 0.252462 1.500571
f 1.053202 -2.338595 -0.374279
g NaN NaN NaN
h -2.359958 -1.157886 -0.551865
In [947]: df.fillna(method='pad', limit=1)
Out[947]:
one two three
a 0.059117 1.138469 -2.400634
b 0.059117 1.138469 -2.400634
c NaN NaN NaN
d NaN NaN NaN
e 0.863937 0.252462 1.500571
f 1.053202 -2.338595 -0.374279
g 1.053202 -2.338595 -0.374279
h -2.359958 -1.157886 -0.551865
To remind you, these are the available filling methods:
| Method | Action |
|---|---|
| pad / ffill | Fill values forward |
| bfill / backfill | Fill values backward |
With time series data, using pad/ffill is extremely common so that the “last known value” is available at every time point.
Dropping axis labels with missing data: dropna¶
You may wish to simply exclude labels from a data set which refer to missing data. To do this, use the dropna method:
In [948]: df
Out[948]:
one two three
a 0.059117 1.138469 -2.400634
b NaN 0.000000 0.000000
c NaN 0.000000 0.000000
d NaN 0.000000 0.000000
e 0.863937 0.252462 1.500571
f 1.053202 -2.338595 -0.374279
g NaN 0.000000 0.000000
h -2.359958 -1.157886 -0.551865
In [949]: df.dropna(axis=0)
Out[949]:
one two three
a 0.059117 1.138469 -2.400634
e 0.863937 0.252462 1.500571
f 1.053202 -2.338595 -0.374279
h -2.359958 -1.157886 -0.551865
In [950]: df.dropna(axis=1)
Out[950]:
two three
a 1.138469 -2.400634
b 0.000000 0.000000
c 0.000000 0.000000
d 0.000000 0.000000
e 0.252462 1.500571
f -2.338595 -0.374279
g 0.000000 0.000000
h -1.157886 -0.551865
In [951]: df['one'].dropna()
Out[951]:
a 0.059117
e 0.863937
f 1.053202
h -2.359958
Name: one
dropna is presently only implemented for Series and DataFrame, but will be eventually added to Panel. Series.dropna is a simpler method as it only has one axis to consider. DataFrame.dropna has considerably more options, which can be examined in the API.
Interpolation¶
A linear interpolate method has been implemented on Series. The default interpolation assumes equally spaced points.
In [952]: ts.count()
Out[952]: 61
In [953]: ts.head()
Out[953]:
2000-01-31 0.469112
2000-02-29 NaN
2000-03-31 NaN
2000-04-28 NaN
2000-05-31 NaN
Freq: BM
In [954]: ts.interpolate().count()
Out[954]: 100
In [955]: ts.interpolate().head()
Out[955]:
2000-01-31 0.469112
2000-02-29 0.435428
2000-03-31 0.401743
2000-04-28 0.368059
2000-05-31 0.334374
Freq: BM
In [956]: fig = plt.figure()
In [957]: ts.interpolate().plot()
Out[957]: <matplotlib.axes.AxesSubplot at 0x116fd5310>
In [958]: plt.close('all')
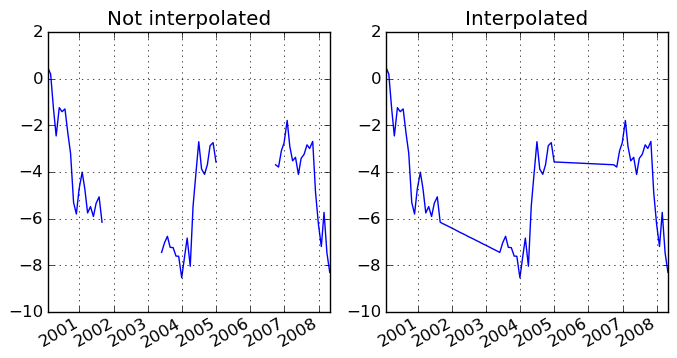
Index aware interpolation is available via the method keyword:
In [959]: ts
Out[959]:
2000-01-31 0.469112
2000-02-29 NaN
2002-07-31 -5.689738
2005-01-31 NaN
2008-04-30 -8.916232
In [960]: ts.interpolate()
Out[960]:
2000-01-31 0.469112
2000-02-29 -2.610313
2002-07-31 -5.689738
2005-01-31 -7.302985
2008-04-30 -8.916232
In [961]: ts.interpolate(method='time')
Out[961]:
2000-01-31 0.469112
2000-02-29 0.273272
2002-07-31 -5.689738
2005-01-31 -7.095568
2008-04-30 -8.916232
For a floating-point index, use method='values':
In [962]: ser
Out[962]:
0 0
1 NaN
10 10
In [963]: ser.interpolate()
Out[963]:
0 0
1 5
10 10
In [964]: ser.interpolate(method='values')
Out[964]:
0 0
1 1
10 10
Replacing Generic Values¶
Often times we want to replace arbitrary values with other values. New in v0.8 is the replace method in Series/DataFrame that provides an efficient yet flexible way to perform such replacements.
For a Series, you can replace a single value or a list of values by another value:
In [965]: ser = Series([0., 1., 2., 3., 4.])
In [966]: ser.replace(0, 5)
Out[966]:
0 5
1 1
2 2
3 3
4 4
You can replace a list of values by a list of other values:
In [967]: ser.replace([0, 1, 2, 3, 4], [4, 3, 2, 1, 0])
Out[967]:
0 4
1 3
2 2
3 1
4 0
You can also specify a mapping dict:
In [968]: ser.replace({0: 10, 1: 100})
Out[968]:
0 10
1 100
2 2
3 3
4 4
For a DataFrame, you can specify individual values by column:
In [969]: df = DataFrame({'a': [0, 1, 2, 3, 4], 'b': [5, 6, 7, 8, 9]})
In [970]: df.replace({'a': 0, 'b': 5}, 100)
Out[970]:
a b
0 100 100
1 1 6
2 2 7
3 3 8
4 4 9
Instead of replacing with specified values, you can treat all given values as missing and interpolate over them:
In [971]: ser.replace([1, 2, 3], method='pad')
Out[971]:
0 0
1 0
2 0
3 0
4 4
Missing data casting rules and indexing¶
While pandas supports storing arrays of integer and boolean type, these types are not capable of storing missing data. Until we can switch to using a native NA type in NumPy, we’ve established some “casting rules” when reindexing will cause missing data to be introduced into, say, a Series or DataFrame. Here they are:
| data type | Cast to |
|---|---|
| integer | float |
| boolean | object |
| float | no cast |
| object | no cast |
For example:
In [972]: s = Series(randn(5), index=[0, 2, 4, 6, 7])
In [973]: s > 0
Out[973]:
0 False
2 True
4 True
6 True
7 True
In [974]: (s > 0).dtype
Out[974]: dtype('bool')
In [975]: crit = (s > 0).reindex(range(8))
In [976]: crit
Out[976]:
0 False
1 NaN
2 True
3 NaN
4 True
5 NaN
6 True
7 True
In [977]: crit.dtype
Out[977]: dtype('object')
Ordinarily NumPy will complain if you try to use an object array (even if it contains boolean values) instead of a boolean array to get or set values from an ndarray (e.g. selecting values based on some criteria). If a boolean vector contains NAs, an exception will be generated:
In [978]: reindexed = s.reindex(range(8)).fillna(0)
In [979]: reindexed[crit]
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
<ipython-input-979-2da204ed1ac7> in <module>()
----> 1 reindexed[crit]
/Users/changshe/code/pandas/pandas/core/series.py in __getitem__(self, key)
447 # special handling of boolean data with NAs stored in object
448 # arrays. Since we can't represent NA with dtype=bool
--> 449 if _is_bool_indexer(key):
450 key = self._check_bool_indexer(key)
451 key = np.asarray(key, dtype=bool)
/Users/changshe/code/pandas/pandas/core/common.py in _is_bool_indexer(key)
499 if not lib.is_bool_array(key):
500 if isnull(key).any():
--> 501 raise ValueError('cannot index with vector containing '
502 'NA / NaN values')
503 return False
ValueError: cannot index with vector containing NA / NaN values
However, these can be filled in using fillna and it will work fine:
In [980]: reindexed[crit.fillna(False)]
Out[980]:
2 1.314232
4 0.690579
6 0.995761
7 2.396780
In [981]: reindexed[crit.fillna(True)]
Out[981]:
1 0.000000
2 1.314232
3 0.000000
4 0.690579
5 0.000000
6 0.995761
7 2.396780