Reshaping and pivot tables#
pandas provides methods for manipulating a Series and DataFrame to alter the
representation of the data for further data processing or data summarization.
pivot()andpivot_table(): Group unique values within one or more discrete categories.stack()andunstack(): Pivot a column or row level to the opposite axis respectively.melt()andwide_to_long(): Unpivot a wideDataFrameto a long format.get_dummies()andfrom_dummies(): Conversions with indicator variables.explode(): Convert a column of list-like values to individual rows.crosstab(): Calculate a cross-tabulation of multiple 1 dimensional factor arrays.cut(): Transform continuous variables to discrete, categorical valuesfactorize(): Encode 1 dimensional variables into integer labels.
pivot() and pivot_table()#
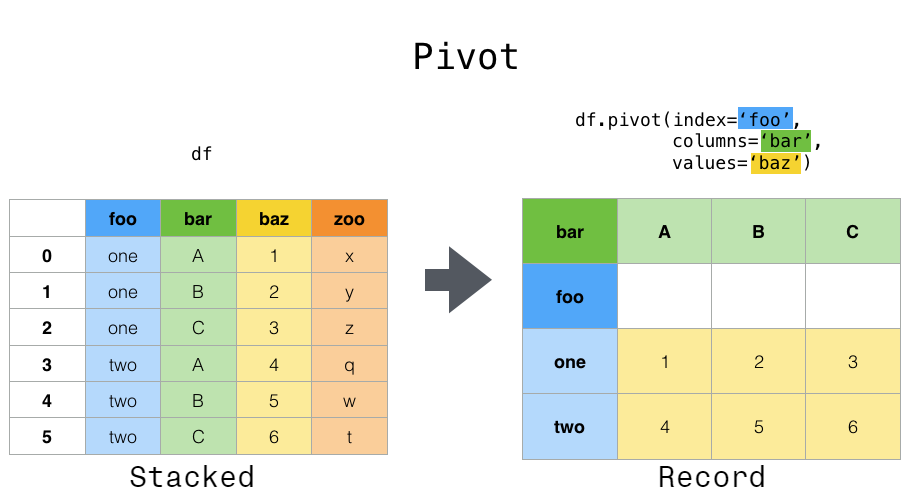
pivot()#
Data is often stored in so-called “stacked” or “record” format. In a “record” or “wide” format, typically there is one row for each subject. In the “stacked” or “long” format there are multiple rows for each subject where applicable.
In [1]: data = {
...: "value": range(12),
...: "variable": ["A"] * 3 + ["B"] * 3 + ["C"] * 3 + ["D"] * 3,
...: "date": pd.to_datetime(["2020-01-03", "2020-01-04", "2020-01-05"] * 4)
...: }
...:
In [2]: df = pd.DataFrame(data)
To perform time series operations with each unique variable, a better
representation would be where the columns are the unique variables and an
index of dates identifies individual observations. To reshape the data into
this form, we use the DataFrame.pivot() method (also implemented as a
top level function pivot()):
In [3]: pivoted = df.pivot(index="date", columns="variable", values="value")
In [4]: pivoted
Out[4]:
variable A B C D
date
2020-01-03 0 3 6 9
2020-01-04 1 4 7 10
2020-01-05 2 5 8 11
If the values argument is omitted, and the input DataFrame has more than
one column of values which are not used as column or index inputs to pivot(),
then the resulting “pivoted” DataFrame will have hierarchical columns whose topmost level indicates the respective value
column:
In [5]: df["value2"] = df["value"] * 2
In [6]: pivoted = df.pivot(index="date", columns="variable")
In [7]: pivoted
Out[7]:
value value2
variable A B C D A B C D
date
2020-01-03 0 3 6 9 0 6 12 18
2020-01-04 1 4 7 10 2 8 14 20
2020-01-05 2 5 8 11 4 10 16 22
You can then select subsets from the pivoted DataFrame:
In [8]: pivoted["value2"]
Out[8]:
variable A B C D
date
2020-01-03 0 6 12 18
2020-01-04 2 8 14 20
2020-01-05 4 10 16 22
Note that this returns a view on the underlying data in the case where the data are homogeneously-typed.
Note
pivot() can only handle unique rows specified by index and columns.
If you data contains duplicates, use pivot_table().
pivot_table()#
While pivot() provides general purpose pivoting with various
data types, pandas also provides pivot_table() or pivot_table()
for pivoting with aggregation of numeric data.
The function pivot_table() can be used to create spreadsheet-style
pivot tables. See the cookbook for some advanced
strategies.
In [9]: import datetime
In [10]: df = pd.DataFrame(
....: {
....: "A": ["one", "one", "two", "three"] * 6,
....: "B": ["A", "B", "C"] * 8,
....: "C": ["foo", "foo", "foo", "bar", "bar", "bar"] * 4,
....: "D": np.random.randn(24),
....: "E": np.random.randn(24),
....: "F": [datetime.datetime(2013, i, 1) for i in range(1, 13)]
....: + [datetime.datetime(2013, i, 15) for i in range(1, 13)],
....: }
....: )
....:
In [11]: df
Out[11]:
A B C D E F
0 one A foo 0.469112 0.404705 2013-01-01
1 one B foo -0.282863 0.577046 2013-02-01
2 two C foo -1.509059 -1.715002 2013-03-01
3 three A bar -1.135632 -1.039268 2013-04-01
4 one B bar 1.212112 -0.370647 2013-05-01
.. ... .. ... ... ... ...
19 three B foo -1.087401 -0.472035 2013-08-15
20 one C foo -0.673690 -0.013960 2013-09-15
21 one A bar 0.113648 -0.362543 2013-10-15
22 two B bar -1.478427 -0.006154 2013-11-15
23 three C bar 0.524988 -0.923061 2013-12-15
[24 rows x 6 columns]
In [12]: pd.pivot_table(df, values="D", index=["A", "B"], columns=["C"])
Out[12]:
C bar foo
A B
one A -0.995460 0.595334
B 0.393570 -0.494817
C 0.196903 -0.767769
three A -0.431886 NaN
B NaN -1.065818
C 0.798396 NaN
two A NaN 0.197720
B -0.986678 NaN
C NaN -1.274317
In [13]: pd.pivot_table(
....: df, values=["D", "E"],
....: index=["B"],
....: columns=["A", "C"],
....: aggfunc="sum",
....: )
....:
Out[13]:
D ... E
A one three ... three two
C bar foo bar ... foo bar foo
B ...
A -1.990921 1.190667 -0.863772 ... NaN NaN -1.067650
B 0.787140 -0.989634 NaN ... 0.372851 1.63741 NaN
C 0.393806 -1.535539 1.596791 ... NaN NaN -3.491906
[3 rows x 12 columns]
In [14]: pd.pivot_table(
....: df, values="E",
....: index=["B", "C"],
....: columns=["A"],
....: aggfunc=["sum", "mean"],
....: )
....:
Out[14]:
sum mean
A one three two one three two
B C
A bar -0.471593 -2.008182 NaN -0.235796 -1.004091 NaN
foo 0.761726 NaN -1.067650 0.380863 NaN -0.533825
B bar -1.665170 NaN 1.637410 -0.832585 NaN 0.818705
foo -0.097554 0.372851 NaN -0.048777 0.186425 NaN
C bar -0.744154 -2.392449 NaN -0.372077 -1.196224 NaN
foo 1.061810 NaN -3.491906 0.530905 NaN -1.745953
The result is a DataFrame potentially having a MultiIndex on the
index or column. If the values column name is not given, the pivot table
will include all of the data in an additional level of hierarchy in the columns:
In [15]: pd.pivot_table(df[["A", "B", "C", "D", "E"]], index=["A", "B"], columns=["C"])
Out[15]:
D E
C bar foo bar foo
A B
one A -0.995460 0.595334 -0.235796 0.380863
B 0.393570 -0.494817 -0.832585 -0.048777
C 0.196903 -0.767769 -0.372077 0.530905
three A -0.431886 NaN -1.004091 NaN
B NaN -1.065818 NaN 0.186425
C 0.798396 NaN -1.196224 NaN
two A NaN 0.197720 NaN -0.533825
B -0.986678 NaN 0.818705 NaN
C NaN -1.274317 NaN -1.745953
Also, you can use Grouper for index and columns keywords. For detail of Grouper, see Grouping with a Grouper specification.
In [16]: pd.pivot_table(df, values="D", index=pd.Grouper(freq="ME", key="F"), columns="C")
Out[16]:
C bar foo
F
2013-01-31 NaN 0.595334
2013-02-28 NaN -0.494817
2013-03-31 NaN -1.274317
2013-04-30 -0.431886 NaN
2013-05-31 0.393570 NaN
2013-06-30 0.196903 NaN
2013-07-31 NaN 0.197720
2013-08-31 NaN -1.065818
2013-09-30 NaN -0.767769
2013-10-31 -0.995460 NaN
2013-11-30 -0.986678 NaN
2013-12-31 0.798396 NaN
Adding margins#
Passing margins=True to pivot_table() will add a row and column with an
All label with partial group aggregates across the categories on the
rows and columns:
In [17]: table = df.pivot_table(
....: index=["A", "B"],
....: columns="C",
....: values=["D", "E"],
....: margins=True,
....: aggfunc="std"
....: )
....:
In [18]: table
Out[18]:
D E
C bar foo All bar foo All
A B
one A 1.568517 0.178504 1.293926 0.179247 0.033718 0.371275
B 1.157593 0.299748 0.860059 0.653280 0.885047 0.779837
C 0.523425 0.133049 0.638297 1.111310 0.770555 0.938819
three A 0.995247 NaN 0.995247 0.049748 NaN 0.049748
B NaN 0.030522 0.030522 NaN 0.931203 0.931203
C 0.386657 NaN 0.386657 0.386312 NaN 0.386312
two A NaN 0.111032 0.111032 NaN 1.146201 1.146201
B 0.695438 NaN 0.695438 1.166526 NaN 1.166526
C NaN 0.331975 0.331975 NaN 0.043771 0.043771
All 1.014073 0.713941 0.871016 0.881376 0.984017 0.923568
Additionally, you can call DataFrame.stack() to display a pivoted DataFrame
as having a multi-level index:
In [19]: table.stack(future_stack=True)
Out[19]:
D E
A B C
one A bar 1.568517 0.179247
foo 0.178504 0.033718
All 1.293926 0.371275
B bar 1.157593 0.653280
foo 0.299748 0.885047
... ... ...
two C foo 0.331975 0.043771
All 0.331975 0.043771
All bar 1.014073 0.881376
foo 0.713941 0.984017
All 0.871016 0.923568
[30 rows x 2 columns]
stack() and unstack()#
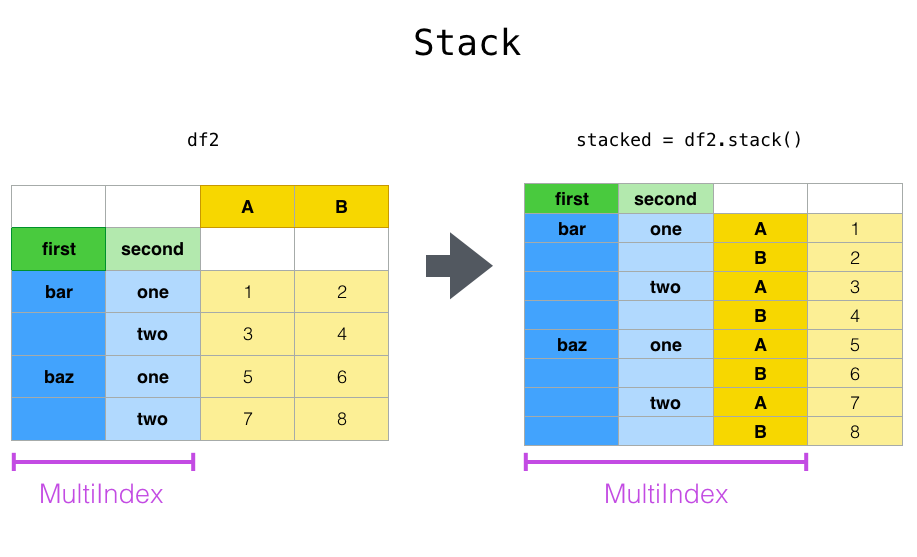
Closely related to the pivot() method are the related
stack() and unstack() methods available on
Series and DataFrame. These methods are designed to work together with
MultiIndex objects (see the section on hierarchical indexing).
stack(): “pivot” a level of the (possibly hierarchical) column labels, returning aDataFramewith an index with a new inner-most level of row labels.unstack(): (inverse operation ofstack()) “pivot” a level of the (possibly hierarchical) row index to the column axis, producing a reshapedDataFramewith a new inner-most level of column labels.
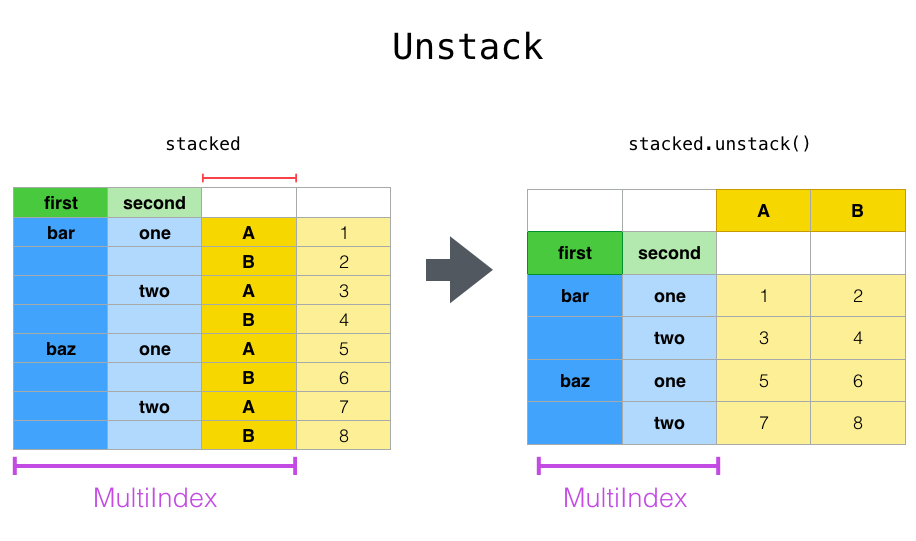
In [20]: tuples = [
....: ["bar", "bar", "baz", "baz", "foo", "foo", "qux", "qux"],
....: ["one", "two", "one", "two", "one", "two", "one", "two"],
....: ]
....:
In [21]: index = pd.MultiIndex.from_arrays(tuples, names=["first", "second"])
In [22]: df = pd.DataFrame(np.random.randn(8, 2), index=index, columns=["A", "B"])
In [23]: df2 = df[:4]
In [24]: df2
Out[24]:
A B
first second
bar one 0.895717 0.805244
two -1.206412 2.565646
baz one 1.431256 1.340309
two -1.170299 -0.226169
The stack() function “compresses” a level in the DataFrame columns to
produce either:
A
DataFrame, in the case of aMultiIndexin the columns.
If the columns have a MultiIndex, you can choose which level to stack. The
stacked level becomes the new lowest level in a MultiIndex on the columns:
In [25]: stacked = df2.stack(future_stack=True)
In [26]: stacked
Out[26]:
first second
bar one A 0.895717
B 0.805244
two A -1.206412
B 2.565646
baz one A 1.431256
B 1.340309
two A -1.170299
B -0.226169
dtype: float64
With a “stacked” DataFrame or Series (having a MultiIndex as the
index), the inverse operation of stack() is unstack(), which by default
unstacks the last level:
In [27]: stacked.unstack()
Out[27]:
A B
first second
bar one 0.895717 0.805244
two -1.206412 2.565646
baz one 1.431256 1.340309
two -1.170299 -0.226169
In [28]: stacked.unstack(1)
Out[28]:
second one two
first
bar A 0.895717 -1.206412
B 0.805244 2.565646
baz A 1.431256 -1.170299
B 1.340309 -0.226169
In [29]: stacked.unstack(0)
Out[29]:
first bar baz
second
one A 0.895717 1.431256
B 0.805244 1.340309
two A -1.206412 -1.170299
B 2.565646 -0.226169
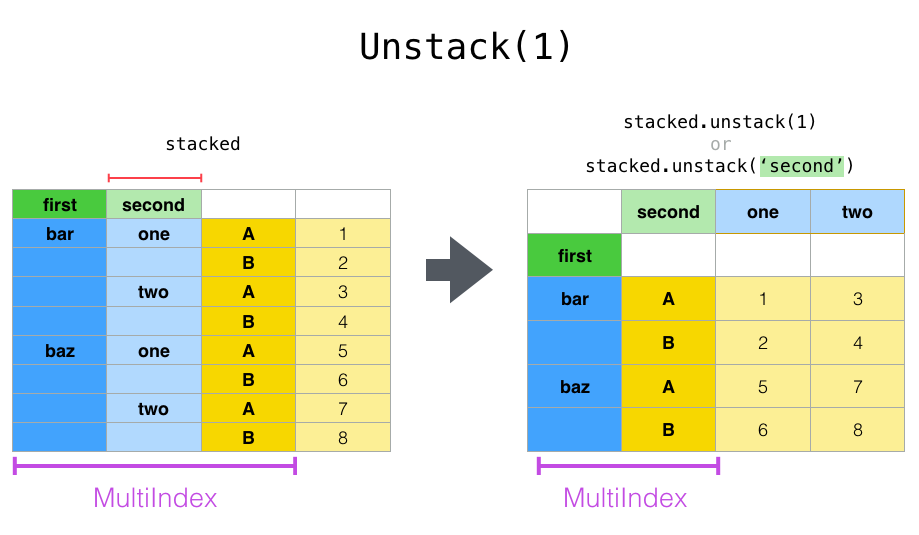
If the indexes have names, you can use the level names instead of specifying the level numbers:
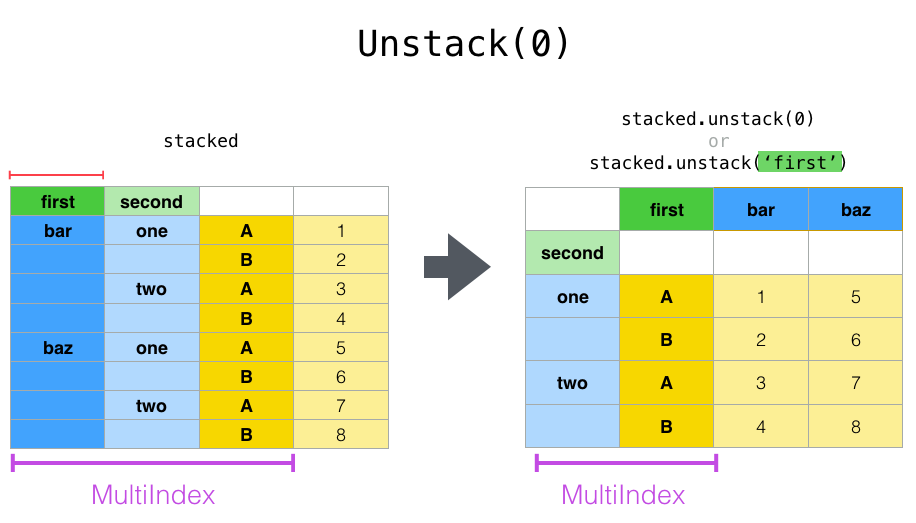
In [30]: stacked.unstack("second")
Out[30]:
second one two
first
bar A 0.895717 -1.206412
B 0.805244 2.565646
baz A 1.431256 -1.170299
B 1.340309 -0.226169
Notice that the stack() and unstack() methods implicitly sort the index
levels involved. Hence a call to stack() and then unstack(), or vice versa,
will result in a sorted copy of the original DataFrame or Series:
In [31]: index = pd.MultiIndex.from_product([[2, 1], ["a", "b"]])
In [32]: df = pd.DataFrame(np.random.randn(4), index=index, columns=["A"])
In [33]: df
Out[33]:
A
2 a -1.413681
b 1.607920
1 a 1.024180
b 0.569605
In [34]: all(df.unstack().stack(future_stack=True) == df.sort_index())
Out[34]: True
Multiple levels#
You may also stack or unstack more than one level at a time by passing a list of levels, in which case the end result is as if each level in the list were processed individually.
In [35]: columns = pd.MultiIndex.from_tuples(
....: [
....: ("A", "cat", "long"),
....: ("B", "cat", "long"),
....: ("A", "dog", "short"),
....: ("B", "dog", "short"),
....: ],
....: names=["exp", "animal", "hair_length"],
....: )
....:
In [36]: df = pd.DataFrame(np.random.randn(4, 4), columns=columns)
In [37]: df
Out[37]:
exp A B A B
animal cat cat dog dog
hair_length long long short short
0 0.875906 -2.211372 0.974466 -2.006747
1 -0.410001 -0.078638 0.545952 -1.219217
2 -1.226825 0.769804 -1.281247 -0.727707
3 -0.121306 -0.097883 0.695775 0.341734
In [38]: df.stack(level=["animal", "hair_length"], future_stack=True)
Out[38]:
exp A B
animal hair_length
0 cat long 0.875906 -2.211372
dog short 0.974466 -2.006747
1 cat long -0.410001 -0.078638
dog short 0.545952 -1.219217
2 cat long -1.226825 0.769804
dog short -1.281247 -0.727707
3 cat long -0.121306 -0.097883
dog short 0.695775 0.341734
The list of levels can contain either level names or level numbers but not a mixture of the two.
# df.stack(level=['animal', 'hair_length'], future_stack=True)
# from above is equivalent to:
In [39]: df.stack(level=[1, 2], future_stack=True)
Out[39]:
exp A B
animal hair_length
0 cat long 0.875906 -2.211372
dog short 0.974466 -2.006747
1 cat long -0.410001 -0.078638
dog short 0.545952 -1.219217
2 cat long -1.226825 0.769804
dog short -1.281247 -0.727707
3 cat long -0.121306 -0.097883
dog short 0.695775 0.341734
Missing data#
Unstacking can result in missing values if subgroups do not have the same set of labels. By default, missing values will be replaced with the default fill value for that data type.
In [40]: columns = pd.MultiIndex.from_tuples(
....: [
....: ("A", "cat"),
....: ("B", "dog"),
....: ("B", "cat"),
....: ("A", "dog"),
....: ],
....: names=["exp", "animal"],
....: )
....:
In [41]: index = pd.MultiIndex.from_product(
....: [("bar", "baz", "foo", "qux"), ("one", "two")], names=["first", "second"]
....: )
....:
In [42]: df = pd.DataFrame(np.random.randn(8, 4), index=index, columns=columns)
In [43]: df3 = df.iloc[[0, 1, 4, 7], [1, 2]]
In [44]: df3
Out[44]:
exp B
animal dog cat
first second
bar one -1.110336 -0.619976
two 0.687738 0.176444
foo one 1.314232 0.690579
qux two 0.380396 0.084844
In [45]: df3.unstack()
Out[45]:
exp B
animal dog cat
second one two one two
first
bar -1.110336 0.687738 -0.619976 0.176444
foo 1.314232 NaN 0.690579 NaN
qux NaN 0.380396 NaN 0.084844
The missing value can be filled with a specific value with the fill_value argument.
In [46]: df3.unstack(fill_value=-1e9)
Out[46]:
exp B
animal dog cat
second one two one two
first
bar -1.110336e+00 6.877384e-01 -6.199759e-01 1.764443e-01
foo 1.314232e+00 -1.000000e+09 6.905793e-01 -1.000000e+09
qux -1.000000e+09 3.803956e-01 -1.000000e+09 8.484421e-02
melt() and wide_to_long()#
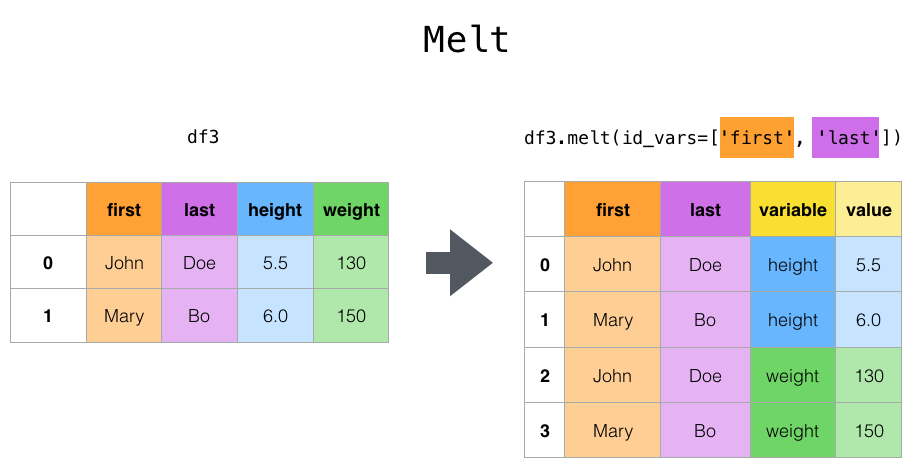
The top-level melt() function and the corresponding DataFrame.melt()
are useful to massage a DataFrame into a format where one or more columns
are identifier variables, while all other columns, considered measured
variables, are “unpivoted” to the row axis, leaving just two non-identifier
columns, “variable” and “value”. The names of those columns can be customized
by supplying the var_name and value_name parameters.
In [47]: cheese = pd.DataFrame(
....: {
....: "first": ["John", "Mary"],
....: "last": ["Doe", "Bo"],
....: "height": [5.5, 6.0],
....: "weight": [130, 150],
....: }
....: )
....:
In [48]: cheese
Out[48]:
first last height weight
0 John Doe 5.5 130
1 Mary Bo 6.0 150
In [49]: cheese.melt(id_vars=["first", "last"])
Out[49]:
first last variable value
0 John Doe height 5.5
1 Mary Bo height 6.0
2 John Doe weight 130.0
3 Mary Bo weight 150.0
In [50]: cheese.melt(id_vars=["first", "last"], var_name="quantity")
Out[50]:
first last quantity value
0 John Doe height 5.5
1 Mary Bo height 6.0
2 John Doe weight 130.0
3 Mary Bo weight 150.0
When transforming a DataFrame using melt(), the index will be ignored.
The original index values can be kept by setting the ignore_index=False parameter to False (default is True).
ignore_index=False will however duplicate index values.
In [51]: index = pd.MultiIndex.from_tuples([("person", "A"), ("person", "B")])
In [52]: cheese = pd.DataFrame(
....: {
....: "first": ["John", "Mary"],
....: "last": ["Doe", "Bo"],
....: "height": [5.5, 6.0],
....: "weight": [130, 150],
....: },
....: index=index,
....: )
....:
In [53]: cheese
Out[53]:
first last height weight
person A John Doe 5.5 130
B Mary Bo 6.0 150
In [54]: cheese.melt(id_vars=["first", "last"])
Out[54]:
first last variable value
0 John Doe height 5.5
1 Mary Bo height 6.0
2 John Doe weight 130.0
3 Mary Bo weight 150.0
In [55]: cheese.melt(id_vars=["first", "last"], ignore_index=False)
Out[55]:
first last variable value
person A John Doe height 5.5
B Mary Bo height 6.0
A John Doe weight 130.0
B Mary Bo weight 150.0
wide_to_long() is similar to melt() with more customization for
column matching.
In [56]: dft = pd.DataFrame(
....: {
....: "A1970": {0: "a", 1: "b", 2: "c"},
....: "A1980": {0: "d", 1: "e", 2: "f"},
....: "B1970": {0: 2.5, 1: 1.2, 2: 0.7},
....: "B1980": {0: 3.2, 1: 1.3, 2: 0.1},
....: "X": dict(zip(range(3), np.random.randn(3))),
....: }
....: )
....:
In [57]: dft["id"] = dft.index
In [58]: dft
Out[58]:
A1970 A1980 B1970 B1980 X id
0 a d 2.5 3.2 1.519970 0
1 b e 1.2 1.3 -0.493662 1
2 c f 0.7 0.1 0.600178 2
In [59]: pd.wide_to_long(dft, ["A", "B"], i="id", j="year")
Out[59]:
X A B
id year
0 1970 1.519970 a 2.5
1 1970 -0.493662 b 1.2
2 1970 0.600178 c 0.7
0 1980 1.519970 d 3.2
1 1980 -0.493662 e 1.3
2 1980 0.600178 f 0.1
get_dummies() and from_dummies()#
To convert categorical variables of a Series into a “dummy” or “indicator”,
get_dummies() creates a new DataFrame with columns of the unique
variables and the values representing the presence of those variables per row.
In [60]: df = pd.DataFrame({"key": list("bbacab"), "data1": range(6)})
In [61]: pd.get_dummies(df["key"])
Out[61]:
a b c
0 False True False
1 False True False
2 True False False
3 False False True
4 True False False
5 False True False
In [62]: df["key"].str.get_dummies()
Out[62]:
a b c
0 0 1 0
1 0 1 0
2 1 0 0
3 0 0 1
4 1 0 0
5 0 1 0
prefix adds a prefix to the the column names which is useful for merging the result
with the original DataFrame:
In [63]: dummies = pd.get_dummies(df["key"], prefix="key")
In [64]: dummies
Out[64]:
key_a key_b key_c
0 False True False
1 False True False
2 True False False
3 False False True
4 True False False
5 False True False
In [65]: df[["data1"]].join(dummies)
Out[65]:
data1 key_a key_b key_c
0 0 False True False
1 1 False True False
2 2 True False False
3 3 False False True
4 4 True False False
5 5 False True False
This function is often used along with discretization functions like cut():
In [66]: values = np.random.randn(10)
In [67]: values
Out[67]:
array([ 0.2742, 0.1329, -0.0237, 2.4102, 1.4505, 0.2061, -0.2519,
-2.2136, 1.0633, 1.2661])
In [68]: bins = [0, 0.2, 0.4, 0.6, 0.8, 1]
In [69]: pd.get_dummies(pd.cut(values, bins))
Out[69]:
(0.0, 0.2] (0.2, 0.4] (0.4, 0.6] (0.6, 0.8] (0.8, 1.0]
0 False True False False False
1 True False False False False
2 False False False False False
3 False False False False False
4 False False False False False
5 False True False False False
6 False False False False False
7 False False False False False
8 False False False False False
9 False False False False False
get_dummies() also accepts a DataFrame. By default, object, string,
or categorical type columns are encoded as dummy variables with other columns unaltered.
In [70]: df = pd.DataFrame({"A": ["a", "b", "a"], "B": ["c", "c", "b"], "C": [1, 2, 3]})
In [71]: pd.get_dummies(df)
Out[71]:
C A_a A_b B_b B_c
0 1 True False False True
1 2 False True False True
2 3 True False True False
Specifying the columns keyword will encode a column of any type.
In [72]: pd.get_dummies(df, columns=["A"])
Out[72]:
B C A_a A_b
0 c 1 True False
1 c 2 False True
2 b 3 True False
As with the Series version, you can pass values for the prefix and
prefix_sep. By default the column name is used as the prefix and _ as
the prefix separator. You can specify prefix and prefix_sep in 3 ways:
string: Use the same value for
prefixorprefix_sepfor each column to be encoded.list: Must be the same length as the number of columns being encoded.
dict: Mapping column name to prefix.
In [73]: simple = pd.get_dummies(df, prefix="new_prefix")
In [74]: simple
Out[74]:
C new_prefix_a new_prefix_b new_prefix_b new_prefix_c
0 1 True False False True
1 2 False True False True
2 3 True False True False
In [75]: from_list = pd.get_dummies(df, prefix=["from_A", "from_B"])
In [76]: from_list
Out[76]:
C from_A_a from_A_b from_B_b from_B_c
0 1 True False False True
1 2 False True False True
2 3 True False True False
In [77]: from_dict = pd.get_dummies(df, prefix={"B": "from_B", "A": "from_A"})
In [78]: from_dict
Out[78]:
C from_A_a from_A_b from_B_b from_B_c
0 1 True False False True
1 2 False True False True
2 3 True False True False
To avoid collinearity when feeding the result to statistical models,
specify drop_first=True.
In [79]: s = pd.Series(list("abcaa"))
In [80]: pd.get_dummies(s)
Out[80]:
a b c
0 True False False
1 False True False
2 False False True
3 True False False
4 True False False
In [81]: pd.get_dummies(s, drop_first=True)
Out[81]:
b c
0 False False
1 True False
2 False True
3 False False
4 False False
When a column contains only one level, it will be omitted in the result.
In [82]: df = pd.DataFrame({"A": list("aaaaa"), "B": list("ababc")})
In [83]: pd.get_dummies(df)
Out[83]:
A_a B_a B_b B_c
0 True True False False
1 True False True False
2 True True False False
3 True False True False
4 True False False True
In [84]: pd.get_dummies(df, drop_first=True)
Out[84]:
B_b B_c
0 False False
1 True False
2 False False
3 True False
4 False True
The values can be cast to a different type using the dtype argument.
In [85]: df = pd.DataFrame({"A": list("abc"), "B": [1.1, 2.2, 3.3]})
In [86]: pd.get_dummies(df, dtype=np.float32).dtypes
Out[86]:
B float64
A_a float32
A_b float32
A_c float32
dtype: object
New in version 1.5.0.
from_dummies() converts the output of get_dummies() back into
a Series of categorical values from indicator values.
In [87]: df = pd.DataFrame({"prefix_a": [0, 1, 0], "prefix_b": [1, 0, 1]})
In [88]: df
Out[88]:
prefix_a prefix_b
0 0 1
1 1 0
2 0 1
In [89]: pd.from_dummies(df, sep="_")
Out[89]:
prefix
0 b
1 a
2 b
Dummy coded data only requires k - 1 categories to be included, in this case
the last category is the default category. The default category can be modified with
default_category.
In [90]: df = pd.DataFrame({"prefix_a": [0, 1, 0]})
In [91]: df
Out[91]:
prefix_a
0 0
1 1
2 0
In [92]: pd.from_dummies(df, sep="_", default_category="b")
Out[92]:
prefix
0 b
1 a
2 b
explode()#
For a DataFrame column with nested, list-like values, explode() will transform
each list-like value to a separate row. The resulting Index will be duplicated corresponding
to the index label from the original row:
In [93]: keys = ["panda1", "panda2", "panda3"]
In [94]: values = [["eats", "shoots"], ["shoots", "leaves"], ["eats", "leaves"]]
In [95]: df = pd.DataFrame({"keys": keys, "values": values})
In [96]: df
Out[96]:
keys values
0 panda1 [eats, shoots]
1 panda2 [shoots, leaves]
2 panda3 [eats, leaves]
In [97]: df["values"].explode()
Out[97]:
0 eats
0 shoots
1 shoots
1 leaves
2 eats
2 leaves
Name: values, dtype: object
DataFrame.explode can also explode the column in the DataFrame.
In [98]: df.explode("values")
Out[98]:
keys values
0 panda1 eats
0 panda1 shoots
1 panda2 shoots
1 panda2 leaves
2 panda3 eats
2 panda3 leaves
Series.explode() will replace empty lists with a missing value indicator and preserve scalar entries.
In [99]: s = pd.Series([[1, 2, 3], "foo", [], ["a", "b"]])
In [100]: s
Out[100]:
0 [1, 2, 3]
1 foo
2 []
3 [a, b]
dtype: object
In [101]: s.explode()
Out[101]:
0 1
0 2
0 3
1 foo
2 NaN
3 a
3 b
dtype: object
A comma-separated string value can be split into individual values in a list and then exploded to a new row.
In [102]: df = pd.DataFrame([{"var1": "a,b,c", "var2": 1}, {"var1": "d,e,f", "var2": 2}])
In [103]: df.assign(var1=df.var1.str.split(",")).explode("var1")
Out[103]:
var1 var2
0 a 1
0 b 1
0 c 1
1 d 2
1 e 2
1 f 2
crosstab()#
Use crosstab() to compute a cross-tabulation of two (or more)
factors. By default crosstab() computes a frequency table of the factors
unless an array of values and an aggregation function are passed.
Any Series passed will have their name attributes used unless row or column
names for the cross-tabulation are specified
In [104]: a = np.array(["foo", "foo", "bar", "bar", "foo", "foo"], dtype=object)
In [105]: b = np.array(["one", "one", "two", "one", "two", "one"], dtype=object)
In [106]: c = np.array(["dull", "dull", "shiny", "dull", "dull", "shiny"], dtype=object)
In [107]: pd.crosstab(a, [b, c], rownames=["a"], colnames=["b", "c"])
Out[107]:
b one two
c dull shiny dull shiny
a
bar 1 0 0 1
foo 2 1 1 0
If crosstab() receives only two Series, it will provide a frequency table.
In [108]: df = pd.DataFrame(
.....: {"A": [1, 2, 2, 2, 2], "B": [3, 3, 4, 4, 4], "C": [1, 1, np.nan, 1, 1]}
.....: )
.....:
In [109]: df
Out[109]:
A B C
0 1 3 1.0
1 2 3 1.0
2 2 4 NaN
3 2 4 1.0
4 2 4 1.0
In [110]: pd.crosstab(df["A"], df["B"])
Out[110]:
B 3 4
A
1 1 0
2 1 3
crosstab() can also summarize to Categorical data.
In [111]: foo = pd.Categorical(["a", "b"], categories=["a", "b", "c"])
In [112]: bar = pd.Categorical(["d", "e"], categories=["d", "e", "f"])
In [113]: pd.crosstab(foo, bar)
Out[113]:
col_0 d e
row_0
a 1 0
b 0 1
For Categorical data, to include all of data categories even if the actual data does
not contain any instances of a particular category, use dropna=False.
In [114]: pd.crosstab(foo, bar, dropna=False)
Out[114]:
col_0 d e f
row_0
a 1 0 0
b 0 1 0
c 0 0 0
Normalization#
Frequency tables can also be normalized to show percentages rather than counts
using the normalize argument:
In [115]: pd.crosstab(df["A"], df["B"], normalize=True)
Out[115]:
B 3 4
A
1 0.2 0.0
2 0.2 0.6
normalize can also normalize values within each row or within each column:
In [116]: pd.crosstab(df["A"], df["B"], normalize="columns")
Out[116]:
B 3 4
A
1 0.5 0.0
2 0.5 1.0
crosstab() can also accept a third Series and an aggregation function
(aggfunc) that will be applied to the values of the third Series within
each group defined by the first two Series:
In [117]: pd.crosstab(df["A"], df["B"], values=df["C"], aggfunc="sum")
Out[117]:
B 3 4
A
1 1.0 NaN
2 1.0 2.0
Adding margins#
margins=True will add a row and column with an All label with partial group aggregates
across the categories on the rows and columns:
In [118]: pd.crosstab(
.....: df["A"], df["B"], values=df["C"], aggfunc="sum", normalize=True, margins=True
.....: )
.....:
Out[118]:
B 3 4 All
A
1 0.25 0.0 0.25
2 0.25 0.5 0.75
All 0.50 0.5 1.00
cut()#
The cut() function computes groupings for the values of the input
array and is often used to transform continuous variables to discrete or
categorical variables:
An integer bins will form equal-width bins.
In [119]: ages = np.array([10, 15, 13, 12, 23, 25, 28, 59, 60])
In [120]: pd.cut(ages, bins=3)
Out[120]:
[(9.95, 26.667], (9.95, 26.667], (9.95, 26.667], (9.95, 26.667], (9.95, 26.667], (9.95, 26.667], (26.667, 43.333], (43.333, 60.0], (43.333, 60.0]]
Categories (3, interval[float64, right]): [(9.95, 26.667] < (26.667, 43.333] < (43.333, 60.0]]
A list of ordered bin edges will assign an interval for each variable.
In [121]: pd.cut(ages, bins=[0, 18, 35, 70])
Out[121]:
[(0, 18], (0, 18], (0, 18], (0, 18], (18, 35], (18, 35], (18, 35], (35, 70], (35, 70]]
Categories (3, interval[int64, right]): [(0, 18] < (18, 35] < (35, 70]]
If the bins keyword is an IntervalIndex, then these will be
used to bin the passed data.
In [122]: pd.cut(ages, bins=pd.IntervalIndex.from_breaks([0, 40, 70]))
Out[122]:
[(0, 40], (0, 40], (0, 40], (0, 40], (0, 40], (0, 40], (0, 40], (40, 70], (40, 70]]
Categories (2, interval[int64, right]): [(0, 40] < (40, 70]]
factorize()#
factorize() encodes 1 dimensional values into integer labels. Missing values
are encoded as -1.
In [123]: x = pd.Series(["A", "A", np.nan, "B", 3.14, np.inf])
In [124]: x
Out[124]:
0 A
1 A
2 NaN
3 B
4 3.14
5 inf
dtype: object
In [125]: labels, uniques = pd.factorize(x)
In [126]: labels
Out[126]: array([ 0, 0, -1, 1, 2, 3])
In [127]: uniques
Out[127]: Index(['A', 'B', 3.14, inf], dtype='object')
Categorical will similarly encode 1 dimensional values for further
categorical operations
In [128]: pd.Categorical(x)
Out[128]:
['A', 'A', NaN, 'B', 3.14, inf]
Categories (4, object): [3.14, inf, 'A', 'B']